The pathogenesis of mixed-lineage leukemia - PubMed (original) (raw)
Review
The pathogenesis of mixed-lineage leukemia
Andrew G Muntean et al. Annu Rev Pathol. 2012.
Abstract
Aggressive leukemias arise in both children and adults as a result of rearrangements to the mixed-lineage leukemia gene (MLL) located on chromosome 11q23. MLL encodes a large histone methyltransferase that directly binds DNA and positively regulates gene transcription, including homeobox (HOX) genes. MLL is involved in chromosomal translocations, partial tandem duplications, and amplifications, all of which result in hematopoietic malignancies due to sustained HOX expression and stalled differentiation. MLL lesions are associated with both acute myeloid leukemia and acute lymphoid leukemia and are usually associated with a relatively poor prognosis despite improved treatment options such as allogeneic hematopoietic stem cell transplantation, which underscores the need for new treatment regimens. Recent advances have begun to reveal the molecular mechanisms that drive MLL-associated leukemias, which, in turn, have provided opportunities for therapeutic development. Here, we discuss the etiology of MLL leukemias and potential directions for future therapy.
Figures
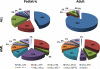
Figure 1
Translocations with MLL occur with a large and diverse group of partner genes. The frequency of some of the most common translocations with MLL are shown for both pediatric and adult ALL or AML. In general, MLL is fused with a more diverse group of partner proteins in AML compared to ALL, which is primarily composed of AF4, AF9 and ENL translocations (6).
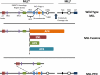
Figure 2
Structure of wild type and leukemia associated MLL proteins. Top: Domain architecture of wild type MLL. MLL is a large multi-domain protein of about 4000 amino acids. Cleavage of MLL (denoted by the blue arrow) results in 320 kDa MLLN and 180 kDa MLLC fragments that non-covalently associate. Domains within MLLN include three AT hooks (red), two subnuclear localization motifs (SNL) (purple), a DNMT1 homology region (CxxC) (Green), four plant homeodomain (PHD) fingers (blue), an atypical bromodomain (orange) and a FYRN domain (open circle). The Breakpoint Cluster Region (BCR) spans an 8.3 kb region bound by BamH1 restriction sites and encompasses exons 5-11 or 7-13 using old or new nomenclature respectively and is the site of chromosomal translocations involving MLL. Between the CxxC and first PHD finger is repression domain 2 (RD2) that is rich in basic amino acids. A HCF binding motif (HBM) (yellow) is found between the bromodomain and PHD3. MLLC contains a transactivation domain (TAD) (filled oval), a FYRC domain (open square) and C-terminal SET domain (dark blue). Middle: Chromosomal translocations involving MLL result in chimeric MLL fusion proteins that include N-terminal sequence of MLL up to the BCR (dotted vertical line) followed by one of several different fusion partners. Examples of fusion partner proteins including AF4, ENL, AF9 and AF6 are shown. MLL fusion proteins invariably retain AT-hooks, SNL1/2 and the CxxC domain of MLLN while losing the downstream PHD fingers and beyond. Bottom: The MLL gene is also prone to internal tandem duplications (MLL-PTD) resulting in duplication of MLL sequences comprising the AT-hooks, SNL1/2 and the CxxC domain which are inserted at the BCR.
Figure 3
MLL interacting proteins. A) MLL is known to interact with a variety of partner proteins. Sequences at the extreme N-terminus of MLL are necessary for interaction with Menin and LEDGF. Together these proteins form a trimeric complex that is necessary for leukemogenesis. PAFc interacts with sequences of MLL retained in MLL fusion proteins and is also critically required for MLL fusion leukemogenesis. The RD2 region of MLL interacts with the co-repressors CtBP, the PcG proteins HPC2 and BMI-1, and the histone deacetylase HDAC1 and appears to be regulated by the binding of the cyclophilin Cyp33 to the third PHD finger of MLL. Host Cell Factor 1 and 2 (HCF1/2) interacts with a HBM consensus sequence found between the bromodomain and PHD4 and links MLL to the function of E2F proteins. The transactivation domain of MLL recruits the HAT CBP, which promotes histone acetylation and gene transcription. MOF associates with MLLC and delivers H4K16 acetyltransferase activity to MLL target genes. MLLC also associates with a core complex of proteins including RbBP5, WDR5 and ASH2L, which are necessary for MLL methyltransferase activity. B) Two Menin Binding Motifs (MBM) with varied binding affinities are found at the N-terminus of MLL and necessary for proper interactions with Menin. C) MLL associates with the PAF complex through two interaction sites. The crystal structure of the MLL CxxC domain shows how the pre and post CxxC domain are in close proximity because of the hairpin folding of the CxxC domain. This likely creates a single binding surface of MLL for making direct interactions with the PAF1 and CTR9 components of PAFc.
Figure 4
MLL complex proteins during normal and malignant hematopoiesis. MLL interacts with a variety of protein complexes in hematopoietic stem and progenitor cells to promote transcription of critical target genes like HOXA9 and MEIS1. The PAF complex associates with RNA pol II and recruits the RAD6/BRE1 E2/E3 ubiquitin ligase, which promotes mono-ubiquitination of histone H2B (Ub). H2B mono-ubiquitination is a histone mark associated with transcriptional activation. PAFc, along with Menin/LEDGF, recruit the MLL complex to target genes which delivers H3K4 (Me) methyltransferase activity and promotes gene transcription. MLL associates with the HAT MOF, which promotes further gene transcription through histone H4K16 acetylation (Ac). During hematopoietic differentiation, MLL is not recruited to target genes in part due to decreased transcription of PAFc. Insufficient recruitment of MLL leads to decreased expression of target genes. Chromosomal translocations involving MLL generate MLL fusion proteins that can recruit transcriptional activation complexes dependent on the fusion partner. MLL translocation partners including AF4, AF9, ENL, ELL and AF5q31 can form a transcriptional activation complex and a related complex AEP (
A
F4/
E
NL/
P
-TEFb) has also been reported. These complexes involve the recruitment of pTEFb, which is required to phosphorylate the RNA Pol II C terminal domain, which promotes transcriptional elongation. The H3K79 methyltransferase DOT1l is also recruited to some MLL fusion proteins (MLL-AF10), which can further promote transcriptional activation.
Similar articles
- The molecular biology of mixed lineage leukemia.
Slany RK. Slany RK. Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review. - _MLL_-Rearranged Acute Leukemia with t(4;11)(q21;q23)-Current Treatment Options. Is There a Role for CAR-T Cell Therapy?
Britten O, Ragusa D, Tosi S, Kamel YM. Britten O, et al. Cells. 2019 Oct 29;8(11):1341. doi: 10.3390/cells8111341. Cells. 2019. PMID: 31671855 Free PMC article. Review. - Monitoring of post-transplant MLL-PTD as minimal residual disease can predict relapse after allogeneic HSCT in patients with acute myeloid leukemia and myelodysplastic syndrome.
Kong J, Gao MG, Qin YZ, Wang Y, Yan CH, Sun YQ, Chang YJ, Xu LP, Zhang XH, Liu KY, Huang XJ, Zhao XS. Kong J, et al. BMC Cancer. 2022 Jan 3;22(1):11. doi: 10.1186/s12885-021-09051-5. BMC Cancer. 2022. PMID: 34979982 Free PMC article. - The MLL gene and translocations involving chromosomal band 11q23 in acute leukemia.
De Braekeleer M, Morel F, Le Bris MJ, Herry A, Douet-Guilbert N. De Braekeleer M, et al. Anticancer Res. 2005 May-Jun;25(3B):1931-44. Anticancer Res. 2005. PMID: 16158928 Review. - MLL: a histone methyltransferase disrupted in leukemia.
Hess JL. Hess JL. Trends Mol Med. 2004 Oct;10(10):500-7. doi: 10.1016/j.molmed.2004.08.005. Trends Mol Med. 2004. PMID: 15464450 Review.
Cited by
- Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs.
Svoboda LK, Harris A, Bailey NJ, Schwentner R, Tomazou E, von Levetzow C, Magnuson B, Ljungman M, Kovar H, Lawlor ER. Svoboda LK, et al. Epigenetics. 2014 Dec;9(12):1613-25. doi: 10.4161/15592294.2014.988048. Epigenetics. 2014. PMID: 25625846 Free PMC article. - The RS4;11 cell line as a model for leukaemia with t(4;11)(q21;q23): Revised characterisation of cytogenetic features.
Ragusa D, Makarov EM, Britten O, Moralli D, Green CM, Tosi S. Ragusa D, et al. Cancer Rep (Hoboken). 2019 Oct;2(5):e1207. doi: 10.1002/cnr2.1207. Epub 2019 Aug 7. Cancer Rep (Hoboken). 2019. PMID: 32721124 Free PMC article. - FDA-approved disulfiram as a novel treatment for aggressive leukemia.
Karsa M, Xiao L, Ronca E, Bongers A, Spurling D, Karsa A, Cantilena S, Mariana A, Failes TW, Arndt GM, Cheung LC, Kotecha RS, Sutton R, Lock RB, Williams O, de Boer J, Haber M, Norris MD, Henderson MJ, Somers K. Karsa M, et al. J Mol Med (Berl). 2024 Apr;102(4):507-519. doi: 10.1007/s00109-023-02414-4. Epub 2024 Feb 13. J Mol Med (Berl). 2024. PMID: 38349407 Free PMC article. - WDR5 high expression and its effect on tumorigenesis in leukemia.
Ge Z, Song EJ, Kawasawa YI, Li J, Dovat S, Song C. Ge Z, et al. Oncotarget. 2016 Jun 21;7(25):37740-37754. doi: 10.18632/oncotarget.9312. Oncotarget. 2016. PMID: 27192115 Free PMC article. - Disordered epigenetic regulation in MLL-related leukemia.
Zhang Y, Chen A, Yan XM, Huang G. Zhang Y, et al. Int J Hematol. 2012 Oct;96(4):428-37. doi: 10.1007/s12185-012-1180-0. Epub 2012 Sep 29. Int J Hematol. 2012. PMID: 23054645 Review.
References
- Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1992;2:113–8. - PubMed
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–8. - PubMed
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. - PubMed
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. - PubMed
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials