The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation - PubMed (original) (raw)
. 2011 Oct 21;44(2):252-64.
doi: 10.1016/j.molcel.2011.09.010.
Esteban Toro, Matthew A Wright, Gregory J Porreca, Davide Baù, Sun-Hae Hong, Michael J Fero, Lihua J Zhu, Marc A Marti-Renom, Harley H McAdams, Lucy Shapiro, Job Dekker, George M Church
Affiliations
- PMID: 22017872
- PMCID: PMC3874842
- DOI: 10.1016/j.molcel.2011.09.010
The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation
Mark A Umbarger et al. Mol Cell. 2011.
Abstract
We have determined the three-dimensional (3D) architecture of the Caulobacter crescentus genome by combining genome-wide chromatin interaction detection, live-cell imaging, and computational modeling. Using chromosome conformation capture carbon copy (5C), we derive ~13 kb resolution 3D models of the Caulobacter genome. The resulting models illustrate that the genome is ellipsoidal with periodically arranged arms. The parS sites, a pair of short contiguous sequence elements known to be involved in chromosome segregation, are positioned at one pole, where they anchor the chromosome to the cell and contribute to the formation of a compact chromatin conformation. Repositioning these elements resulted in rotations of the chromosome that changed the subcellular positions of most genes. Such rotations did not lead to large-scale changes in gene expression, indicating that genome folding does not strongly affect gene regulation. Collectively, our data suggest that genome folding is globally dictated by the parS sites and chromosome segregation.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Figure 1. Genome-wide 5C reveals that the swarmer chromosome is ellipsoidal
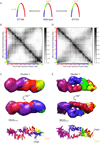
(A) Outline of our 3C and 5C approaches. (Top) Formaldehyde is added to a culture of synchronized swarmer cells, cross-linking physically touching chromosomal loci (e.g. the red and purple regions denoted within the dashed circle). Chromatin is subsequently digested with a restriction enzyme and is diluted in order to separate non-cross-linked fragments. Ligation is then performed, yielding a 3C library of ligation products in which the original inter-fragment cross-links are now represented as ligation products between restriction fragments. (Bottom) Next, bar-coded oligonucleotide probes with sequences complementary to the ends of restriction fragments (plus or minus strand) are annealed to the 3C library. Multiplexed ligation is subsequently performed, creating carbon copy ligation products between annealed plus and minus strand probes that reflect the original cross-links between restriction fragments. By design ligation can only occur between plus and minus strand probes. Carbon copy junctions are subsequently amplified using universal PCR primers that recognize the common tails of the probes, yielding a 5C library, and the frequencies of particular products within this library are assessed via polony-sequencing of barcodes. (B) 3C recapitulates previously established features of Caulobacter swarmer cell genome spatial organization. Template dilution (2-fold) series PCR reactions querying ligation products amongst origin and terminus proximal restriction fragments in swarmer cell 3C libraries generated with (top) or without (bottom) formaldehyde fixation were performed and the resulting products were run on an agarose ethidium bromide stained gel. C denotes a control template in which all junctions were present in equal concentrations (See Supplemental Experimental Procedures). The fragments queried in the origin-origin, terminus-terminus, and origin-terminus reactions were separated by 2,751, 4,633, and 1,970,992 bp, respectively. (C) 5C probe whole-genome coverage map. The central circle represents the Caulobacter genomic map with coloring indicating genome position (from red to blue). The interior and exterior circles represent the genomic locations of restriction fragments queried by plus (grey-exterior) and minus (black-interior) strand 5C probes. (D) Average swarmer cell 5C contact map. The frequency of a junction between a given pair of fragments in the library (contact frequency) is found at the intersection of the row and column corresponding to the fragments. Minus and plus strand probed restriction fragments are ordered according to genome position along the vertical and horizontal axes, respectively. The origin is located at a genome coordinate of 0 and terminus is located at ~2 Mb. The bars underneath and to the left of the two axes indicate genome position. Color within the heat map is indicative of the contact frequency Z-score (See Supplemental Experimental Procedures). (E) (Bottom) The contact frequency profile for a representative fragment located at ~1 Mb. This profile indicates the contact frequencies for a given fragment with all other fragments of opposite type and is identical to a single row of the contact map. The orange dotted line indicates the genome coordinate of this fragment. (Top) Schematic representing the ellipsoidal geometry suggested by the contact frequency profile shown below. See Figure S1 for the corresponding profile and contact map if the chromosome were an unconstrained circle.
Figure 2. Modeling reveals the 3D architecture of the swarmer genome
(A) Outline of our modeling methodology. Restriction fragments were modeled as points connected by springs. The distance derived from the contact frequency between pair of fragments was used (I) to define the equilibrium length of the spring (See Supplemental Experimental Procedures) that connected these fragments (II). The 3D coordinates of all points were randomly initialized (III) and optimization was performed to derive a structure that minimally violates these equilibrium lengths (IV-a). This initialization and optimization procedure was repeated thousands of times to generate an ensemble of structures. The structures were then superimposed and grouped based upon their coordinates, yielding clusters of models in which the 3D coordinates of restriction fragments are structurally very similar (IV-b). (B) 3D Density map representations of the four clusters (ordered based upon the number of models in each cluster) from a wild-type swarmer modeling run. Each queried fragment is represented by a three-dimensional Gaussian that has a correlation coefficient > 0.8 with the space this fragment occupies across all models within the cluster. The positioning of the maximally polar fragment (located ~7 Kb from the parS) elements is indicated in orange. (C) The centroid model of swarmer clusters 1–4. For more information regarding these clusters see Figure S2 and Table S2.
Figure 3. 5C Analyses of strains carrying genomic inversions reveal that the parS elements are critical to defining chromosome orientation
(A) Genomic maps for strains CB15N (wild-type), and inversion strains ET166, and ET163. The inverted region is indicated in yellow and green. (B/D) Contact maps for strain ET166 and ET163 swarmer cells. (C/E) 3D Density map (top) and cluster centroid (bottom) representations for the two largest ET166 and ET163 model clusters (See Figure S3A/C for representations of the two additional clusters for each strain). In the density representation, each queried fragment is represented by a three-dimensional Gaussian with correlation coefficient > 0.8 with the space this fragment occupies across all models within the cluster. The parS elements are located near the junction of the blue and green portions of the ET166 models and near the junction of the yellow and red regions of the ET163 models.
Figure 4. The genomic positions of the parS sites affect the orientation of the entire Caulobacter genome within the cell
(A) Representative images showing the position of parS (yellow), and a lacO array (blue) inserted 2898 Kb from the origin (cyan), in wild-type and the inversion strains, ET166 and ET163. (B–C) Sub-cellular localization of DNA loci bearing lacO insertions visualized with LacI-CFP in swarmer cells with a wild-type (black) or inverted (ET166-green or ET163-red) chromosome. Each point represents an independent measurement of the average cellular position of LacI-CFP in at least 100 cells. Relative cellular position was measured with respect to the old (stalked) cell pole. The dotted vertical lines denote the inversion break points in strains ET166 and ET163. Cartoons to the right illustrate the gross organization and orientation of the chromosomes before and after inversion. Red points labeled A, B, and C provide guides to match the cartoons to the data. See Figure S4 for data for a strain in which the parS sites are located to the right of the origin.
Figure 5. Inter-arm alignments reveal interaction asymmetries in ET166 swarmer cells
(A) (Top) Wild-type cluster 1 swarmer long-axis alignment plot. The genomic distance of each fragment to the most polar fragment (parS elements) is plotted against the genomic distance of the closest opposite arm fragment (with respect to position along the long axis of the models) to this same maximally polar fragment. The black line indicates a perfectly symmetric pairing and increasingly yellow colors indicate larger deviations from symmetry. Data represents the median for 200 randomly selected models from cluster 1. (Bottom) 3D representation the long-axis alignment in wild-type swarmer cells using the smoothed centroid of the cluster 1. Nearest neighbor opposite arm fragments are connected by lines whose colors indicate the degree of deviation from symmetry. (B) (Top) ET166 cluster 1 long axis alignment plot. (Bottom) 3D representation of the ET166 alignment utilizing the smoothed centroid model from cluster 1. A similar plot for ET163 can be found in Figure S5A.
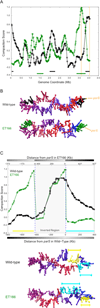
Figure 6. The parS sites nucleate a compact region of the genome
(A) (Top) Wild-type (black) and ET166 (green) genome compaction profile generated from the models in cluster 1. This profile presents the local compaction (derived from the 3D distances between neighboring restriction fragments) as a function of genome position (Supplemental Experimental Procedures). The genome coordinates of restriction fragments in ET166 have been adjusted to account for the inversion present in this strain. Dashed orange lines indicate the positions of the parS sites. Green and black bars indicate the regions highlighted below. (B) The centroid models of wild-type and ET166 cluster 1 with color indicating the pole proximal regions that were determined to be particularly compact (see A). (C) (Top) Plot of compaction versus genome position for wild-type (black) and ET166 (green) model clusters. The compaction score is plotted with fragments ordered along the horizontal axis according to their positions in the wild-type genome. Dashed blue lines denote the inversion break points. In the ET166 genome the shaded region is inverted such that the right-most edge of the shaded region lies adjacent to the region to the left of the left dashed blue line. Distances of fragments from the parS elements in the wild-type and ET166 genome are indicated by the horizontal axes at bottom and top, respectively, with the accompanying horizontal colorbars indicating absolute distance from the parS sites in each strain (darker indicates larger distances). Yellow and cyan lines denote genomic regions that are more compact in ET166 and wild-type models, respectively. These regions are highlighted on the centroid models from the wild-type and ET166 model cluster 1 (Bottom).
Figure 7. The Caulobacter chromosome is free to rotate around the long cell axis
(A) (Left) Schematic of a Caulobacter swarmer cell indicating the positions of the new and old poles as well as the dorsal and ventral sides of the cell. Negative and positive signs refer to the convention used by our image analysis software. (Center) Example micrographs of double-labeled Caulobacter swarmer cells showing configurations of the chromosome in which the labeled loci reside on opposite sides of the cell. (Right) Relative positions of the left and right arm markers in three strains marked at different positions in the chromosome. Circles denote the means of three experiments each of which included at least 400 cells. Bars represent 95% confidence intervals of the mean. The dotted line indicates the expected value for a distribution in which loci have no preferential localization along the short axis. (B) Virtual cell showing the distribution of ~200,000 LacI-CFP foci along the short and long axes of the cell. (Left) Markers on the right arm. (Center) Markers on the left arm. (Right) Merge of the two arms. Note that the two arms are equally distributed along the short cell axis.
Comment in
- Bacterial physiology. Seeing Caulobacter in 3D.
Molloy S. Molloy S. Nat Rev Microbiol. 2011 Nov 16;9(12):834-5. doi: 10.1038/nrmicro2703. Nat Rev Microbiol. 2011. PMID: 22085857 No abstract available. - Three-dimensional genetics.
Rusk N. Rusk N. Nat Methods. 2012 Jan;9(1):14-5. doi: 10.1038/nmeth.1843. Nat Methods. 2012. PMID: 22312629 No abstract available.
Similar articles
- A physical approach to segregation and folding of the Caulobacter crescentus genome.
Dame RT, Tark-Dame M, Schiessel H. Dame RT, et al. Mol Microbiol. 2011 Dec;82(6):1311-5. doi: 10.1111/j.1365-2958.2011.07898.x. Epub 2011 Nov 22. Mol Microbiol. 2011. PMID: 22029843 - Caulobacter requires a dedicated mechanism to initiate chromosome segregation.
Toro E, Hong SH, McAdams HH, Shapiro L. Toro E, et al. Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15435-40. doi: 10.1073/pnas.0807448105. Epub 2008 Sep 29. Proc Natl Acad Sci U S A. 2008. PMID: 18824683 Free PMC article. - Chromosome Dynamics in Bacteria: Triggering Replication at the Opposite Location and Segregation in the Opposite Direction.
Meléndez AB, Menikpurage IP, Mera PE. Meléndez AB, et al. mBio. 2019 Jul 30;10(4):e01002-19. doi: 10.1128/mBio.01002-19. mBio. 2019. PMID: 31363028 Free PMC article. - Chromosome conformation capture assays in bacteria.
Umbarger MA. Umbarger MA. Methods. 2012 Nov;58(3):212-20. doi: 10.1016/j.ymeth.2012.06.017. Epub 2012 Jul 6. Methods. 2012. PMID: 22776362 Review. - Linear ordering and dynamic segregation of the bacterial chromosome.
Breier AM, Cozzarelli NR. Breier AM, et al. Proc Natl Acad Sci U S A. 2004 Jun 22;101(25):9175-6. doi: 10.1073/pnas.0403722101. Epub 2004 Jun 15. Proc Natl Acad Sci U S A. 2004. PMID: 15199189 Free PMC article. Review. No abstract available.
Cited by
- A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae.
Val ME, Marbouty M, de Lemos Martins F, Kennedy SP, Kemble H, Bland MJ, Possoz C, Koszul R, Skovgaard O, Mazel D. Val ME, et al. Sci Adv. 2016 Apr 22;2(4):e1501914. doi: 10.1126/sciadv.1501914. eCollection 2016 Apr. Sci Adv. 2016. PMID: 27152358 Free PMC article. - In vivo facilitated diffusion model.
Bauer M, Metzler R. Bauer M, et al. PLoS One. 2013;8(1):e53956. doi: 10.1371/journal.pone.0053956. Epub 2013 Jan 18. PLoS One. 2013. PMID: 23349772 Free PMC article. - Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis.
Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Wang X, et al. Genes Dev. 2015 Aug 1;29(15):1661-75. doi: 10.1101/gad.265876.115. Genes Dev. 2015. PMID: 26253537 Free PMC article. - Polymer modeling of the E. coli genome reveals the involvement of locus positioning and macrodomain structuring for the control of chromosome conformation and segregation.
Junier I, Boccard F, Espéli O. Junier I, et al. Nucleic Acids Res. 2014 Feb;42(3):1461-73. doi: 10.1093/nar/gkt1005. Epub 2013 Nov 4. Nucleic Acids Res. 2014. PMID: 24194594 Free PMC article. - Cell Boundary Confinement Sets the Size and Position of the E. coli Chromosome.
Wu F, Swain P, Kuijpers L, Zheng X, Felter K, Guurink M, Solari J, Jun S, Shimizu TS, Chaudhuri D, Mulder B, Dekker C. Wu F, et al. Curr Biol. 2019 Jul 8;29(13):2131-2144.e4. doi: 10.1016/j.cub.2019.05.015. Epub 2019 May 30. Curr Biol. 2019. PMID: 31155353 Free PMC article.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. - PubMed
- Alley MR, Maddock JR, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG003143/HG/NHGRI NIH HHS/United States
- R01 GM51426/GM/NIGMS NIH HHS/United States
- R01 HG003143/HG/NHGRI NIH HHS/United States
- R01 GM051426/GM/NIGMS NIH HHS/United States
- R24 GM073011-04/GM/NIGMS NIH HHS/United States
- R24 GM073011/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous