Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor - PubMed (original) (raw)
. 2011 Nov 8;108(45):18324-9.
doi: 10.1073/pnas.1112064108. Epub 2011 Oct 21.
Julie Chaix, Aurore Fenis, Yann M Kerdiles, Nadia Yessaad, Ana Reynders, Claude Gregoire, Herve Luche, Sophie Ugolini, Elena Tomasello, Thierry Walzer, Eric Vivier
Affiliations
- PMID: 22021440
- PMCID: PMC3215049
- DOI: 10.1073/pnas.1112064108
Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor
Emilie Narni-Mancinelli et al. Proc Natl Acad Sci U S A. 2011.
Abstract
NKp46 is a cell surface receptor expressed on natural killer (NK) cells, on a minute subset of T cells, and on a population of innate lymphoid cells that produce IL-22 and express the transcription factor retinoid-related orphan receptor (ROR)-γt, referred to as NK cell receptor (NKR)(+)ROR-γt(+) cells. Here we describe Nkp46(iCre) knock-in mice in which the gene encoding the improved Cre (iCre) recombinase was inserted into the Nkp46 locus. This mouse was used to noninvasively trace cells expressing NKp46 in vivo. Fate mapping experiments demonstrated the stable expression of NKp46 on NK cells and allowed a reappraisal of the sequential steps of NK cell maturation. NKp46 genetic tracing also showed that gut NKR(+)ROR-γt(+) and NK cells represent two distinct lineages. In addition, the genetic heterogeneity of liver NK cells was evidenced. Finally, Nkp46(iCre) mice also represent a unique mouse model of conditional mutagenesis specifically in NKp46(+) cells, paving the way for further developments in the biology of NKp46(+) NK, T, and NKR(+)ROR-γt(+) cells.
Conflict of interest statement
Conflict of interest statement: E.V. is a co-founder and shareholder of Innate-Pharma.
Figures
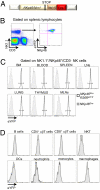
Fig. 1.
Faithful expression of eYFP according to NKp46 expression in _NKp46_iCre_R26R_eYFP mice. (A) Schematic representation of the strategy used to generate _NKp46_iCre mice. (B) Flow cytometric measurement of eYFP and NKp46 expressions on NK cells from spleen of _NKp46_iCre_R26R_eYFP. (C and D) Flow cytometric measurement of eYFP expression on gated NK cells (C) or indicated cell population (D) from indicated organs of _NKp46_iCre_R26R_eYFP (open histograms) and control _NKp46_iCre mice (gray histograms). Results are representative of three experiments.
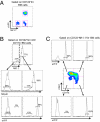
Fig. 2.
Specific depletion of NK cells in DT-treated _NKp46_iCre_R26R_DTR mice. Groups of mice were treated i.v. with DT, and cell recovery was analyzed 2 d later. (A) NK cell proportions among live leukocytes of nontreated (−) or DT-treated mice. (B) Lymphoid cell populations recovery expressed as percent of cell numbers in nontreated mice (mean ± SEM). n ≥ 12 per group and per organ. ***P < 0.001.
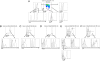
Fig. 3.
Early expression of NKp46 during NK cell maturation. (A) Flow cytometric measurement of NK1.1 and eYFP on gated CD122+lin− NK cells in BM of _NKp46_iCre_R26R_eYFP mice. (B) Flow cytometric measurement of eYFP and NKp46 expression on gated CD122+lin−c-Kit−CD11b−NK1.1− NK precursors and NK1.1+ immature NK cells in BM of _NKp46_iCre_R26R_eYFP mice. (C) eYFP expression is shown on CD122+lin−NK1.1+ BM NK cells according to their expression of CD27 and CD11b markers.
Fig. 4.
CD16 is expressed on immature NK cells. (A) CD16 expression is shown in CD122+lin−NK1.1+ BM NK cells of _NKp46_iCre_R26R_eYFP mice according to CD27 and CD11b expression. eYFP expression is also shown on immature CD27+CD11b−/lowCD16− and CD16+ NK cells. (B_–_F) Flow cytometric measurement of NK cell maturation marker expression (open histograms) or control isotype (shaded histograms) on indicated cells from BM of _NKp46_iCre_R26R_eYFP mice.
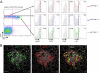
Fig. 5.
Characterization of gut NKp46+eYFP+ and NKp46dim/loweYFPhigh cells. (A) Flow cytometric characterization of CD3-CD19-eYFPhiNK1.1−, eYFPhiNK1.1dim, eYFP+NK1.1+ cells from small intestine of _NKp46_iCre_R26R_eYFP mice for indicated makers (open histograms) or control isotype (gray histograms). Results are representative of two experiments. (B) Immunofluorescence histology of small intestine frozen sections from _NKp46_Cre_R26R_eYFP mice stained with anti-eYFP (green) and anti-RORt (red). Nuclei were counterstained with Sytox (grey). Scale bars, 10 mm. Data are representative of at least three experiments.
Fig. 6.
Characterization of liver NKp46+eYFP+ and NKp46dim/loweYFPhigh cells. Flow cytometric characterization of NK1.1+CD3−eYFPhi and NK1.1+CD3−eYFP+ liver cells from _NKp46_iCre_R26R_eYFP mice for indicated makers (open histograms) or control isotype (gray histograms). Results are representative of two to four experiments.
Similar articles
- IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors.
Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CA, Di Santo JP. Satoh-Takayama N, et al. J Exp Med. 2010 Feb 15;207(2):273-80. doi: 10.1084/jem.20092029. Epub 2010 Feb 8. J Exp Med. 2010. PMID: 20142427 Free PMC article. - Conditional ablation of NKp46+ cells using a novel Ncr1(greenCre) mouse strain: NK cells are essential for protection against pulmonary B16 metastases.
Merzoug LB, Marie S, Satoh-Takayama N, Lesjean S, Albanesi M, Luche H, Fehling HJ, Di Santo JP, Vosshenrich CA. Merzoug LB, et al. Eur J Immunol. 2014 Nov;44(11):3380-91. doi: 10.1002/eji.201444643. Epub 2014 Sep 19. Eur J Immunol. 2014. PMID: 25142413 - The Murine Natural Cytotoxic Receptor NKp46/NCR1 Controls TRAIL Protein Expression in NK Cells and ILC1s.
Sheppard S, Schuster IS, Andoniou CE, Cocita C, Adejumo T, Kung SKP, Sun JC, Degli-Esposti MA, Guerra N. Sheppard S, et al. Cell Rep. 2018 Mar 27;22(13):3385-3392. doi: 10.1016/j.celrep.2018.03.023. Cell Rep. 2018. PMID: 29590608 Free PMC article. - Lessons from NK Cell Deficiencies in the Mouse.
Deauvieau F, Fenis A, Dalençon F, Burdin N, Vivier E, Kerdiles Y. Deauvieau F, et al. Curr Top Microbiol Immunol. 2016;395:173-90. doi: 10.1007/82_2015_473. Curr Top Microbiol Immunol. 2016. PMID: 26385768 Review. - Noncanonical B Cells: Characteristics of Uncharacteristic B Cells.
Haas KM. Haas KM. J Immunol. 2023 Nov 1;211(9):1257-1265. doi: 10.4049/jimmunol.2200944. J Immunol. 2023. PMID: 37844278 Free PMC article. Review.
Cited by
- The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation.
Holmes TD, Pandey RV, Helm EY, Schlums H, Han H, Campbell TM, Drashansky TT, Chiang S, Wu CY, Tao C, Shoukier M, Tolosa E, Von Hardenberg S, Sun M, Klemann C, Marsh RA, Lau CM, Lin Y, Sun JC, Månsson R, Cichocki F, Avram D, Bryceson YT. Holmes TD, et al. Sci Immunol. 2021 Mar 12;6(57):eabc9801. doi: 10.1126/sciimmunol.abc9801. Sci Immunol. 2021. PMID: 33712472 Free PMC article. - Pulmonary Administration of TLR2/6 Agonist after Allergic Sensitization Inhibits Airway Hyper-Responsiveness and Recruits Natural Killer Cells in Lung Parenchyma.
Devulder J, Barrier M, Carrard J, Amniai L, Plé C, Marquillies P, Ledroit V, Ryffel B, Tsicopoulos A, de Nadai P, Duez C. Devulder J, et al. Int J Mol Sci. 2024 Sep 4;25(17):9606. doi: 10.3390/ijms25179606. Int J Mol Sci. 2024. PMID: 39273551 Free PMC article. - Increased CaMKK2 Expression Is an Adaptive Response That Maintains the Fitness of Tumor-Infiltrating Natural Killer Cells.
Juras PK, Racioppi L, Mukherjee D, Artham S, Gao X, Akullian D'Agostino L, Chang CY, McDonnell DP. Juras PK, et al. Cancer Immunol Res. 2023 Jan 3;11(1):109-122. doi: 10.1158/2326-6066.CIR-22-0391. Cancer Immunol Res. 2023. PMID: 36301267 Free PMC article. - Innate lymphoid cell function in the context of adaptive immunity.
Bando JK, Colonna M. Bando JK, et al. Nat Immunol. 2016 Jun 21;17(7):783-9. doi: 10.1038/ni.3484. Nat Immunol. 2016. PMID: 27328008 Free PMC article. - Immune Cell-Type Specific Ablation of Adapter Protein ADAP Differentially Modulates EAE.
Rudolph J, Meinke C, Voss M, Guttek K, Kliche S, Reinhold D, Schraven B, Reinhold A. Rudolph J, et al. Front Immunol. 2019 Oct 1;10:2343. doi: 10.3389/fimmu.2019.02343. eCollection 2019. Front Immunol. 2019. PMID: 31632410 Free PMC article.
References
- Biassoni R, et al. The murine homologue of the human NKp46, a triggering receptor involved in the induction of natural cytotoxicity. Eur J Immunol. 1999;29:1014–1020. - PubMed
- Boysen P, Storset AK. Bovine natural killer cells. Vet Immunol Immunopathol. 2009;130:163–177. - PubMed
- Storset AK, et al. NKp46 defines a subset of bovine leukocytes with natural killer cell characteristics. Eur J Immunol. 2004;34:669–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases