Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene - PubMed (original) (raw)
Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene
Chuan Shen et al. Cancer Discov. 2011 Aug.
Abstract
Kidney cancers often delete chromosome 3p, spanning the VHL tumor suppressor gene, and chromosome 14q, which presumably harbors ≥ 1 tumor suppressor genes. pVHL inhibits the hypoxia-inducible transcription factor (HIF), and HIF2α is a kidney cancer oncoprotein. In this article, we identify focal, homozygous deletions of the HIF1α locus on 14q in clear cell renal carcinoma cell lines. Wild-type HIF1α suppresses renal carcinoma growth, but the products of these altered loci do not. Conversely, downregulation of HIF1α in HIF1α-proficient lines promotes tumor growth. HIF1α activity is diminished in 14q-deleted kidney cancers, and all somatic HIF1α mutations identified in kidney cancers tested to date are loss of function. Therefore, HIF1α has the credentials of a kidney cancer suppressor gene.
Significance: Deletion of 14q is a frequent event in clear cell renal carcinoma and portends a poor prognosis. In this study, we provide genetic and functional evidence that HIF1α is a target of 14q loss in kidney cancer.
Keywords: 14q deletion; HIF1α; hypoxia; kidney cancer; tumor suppression; von Hippel-Lindau.
Conflict of interest statement
Conflict of Interest: W.G.K. owns equity in, and consults for, Fibrogen, Inc., which is developing drugs that modulate HIF activity.
Figures
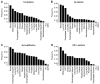
Figure 1. Frequencies of Chromosomal Abnormalities Across Different Cancers
(A) Large deletions affecting most of 14q arm. (B) Large deletions affecting most of 3p arm. (C) Amplification of any region of 5q. (D) Deletions affecting HIF1α locus. See also Figure S1.
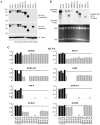
Figure 2. HIF1α Deletions and Altered HIF1α Gene Products in Renal Carcinoma Cells
Immunoblot (A) and Northern blot (B) analysis of the indicated cell lines. The difference in HIF1α electrophoretic mobility between HK-2 immortalized renal epithelial cells and the HIF1α-positive lines such as A704, Caki-2, RCC4, and UMRC-2 might reflect differential phosphorylation. See also Figure S2. (C) Shown are MLPA data for the indicated cell lines, normalized to HK-2 immortalized renal epithelial cells (diploid = 1). Black bars = selected control exons on chromosomes 1, 10, and 17. Grey bars = HIF1α exons. See also Figure S3, Figure S4 and Table S1.
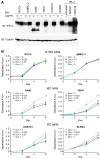
Figure 3. Suppression of _VHL_−/− renal carcinoma proliferation by HIF1α
Immunoblot (A) and proliferation (B) assays of indicated cell lines infected with a doxycycline-inducible retrovirus encoding HIF1α and propagated in 5% serum in the presence or absence of doxycycline. HK-2 cells treated with vehicle, DMOG (1 mM), or MG132 (10 μM) were included as a control in (A). See also Figure S5.
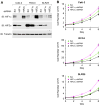
Figure 4. Downregulation of HIF1α in VHL−/− renal carcinoma cells enhances cell proliferation
Immunoblot (A) and proliferation (B) assays of the indicated cell lines after infection with lentiviruses encoding HIF1α shRNA, HIF2α shRNA, or scrambled control shRNA and grown in the presence of 5% serum. See also Figure S6.
Figure 5. Downregulation of HIF1α Promotes Tumor Cell Growth In Vivo
(A) Representative bioluminescent images of nude mice orthotopically injected with RCC4 renal carcinoma cells that stably express firefly luciferase and an shRNA directed against HIF1α (right kidney) or a scrambled shRNA control (left kidney). Shown are images 1 week and 9 week after tumor cell implantation. (B) Bioluminescence ratio as a function of time for mice as in (A). For each mouse the ratio was normalized to the week 1 ratio for that mouse. (C) Mean fold change in normalized bioluminescence ratio for mice analyzed in (B) at week 9. Error bars = 1 standard error of the mean. (D & F) Representative images of nude mice 4 weeks after subcutaneous injection of UMRC-2 renal carcinoma cells that stably express an shRNA directed against HIF1α (right side) or a scrambled shRNA control (left side). (E & G) Mean tumor weights at necropsy of mice as in (D & F). Error bars = 1 standard error of the mean. See also Figure S6.
Figure 6. Differential Effects of Cell Line-Derived HIF1α Variants on Proliferation and Tumor Cell Fitness
(A) Schematic of HIF1α variants identified in renal carcinoma lines. (B and C). Immunoblot (B) and proliferation (C) assays of 769P cells infected with a doxycycline-inducible retroviruses encoding the indicated HIF1α variants and propagated in the presence of doxycyline. 769P infected with an inducible virus encoding wild-type HIF1α but grown in the absence of doxycycline were included as a control in (B). (D) Schematic for in vitro and in vivo competition assays. (E) and (F). PCR-based analysis indicating relative abundance of cells as in (B and C) after growth in vitro for 9 days (E) or orthotopically in vivo for 6 weeks (F) in the presence of absence of doxycycline. Neg. = PCR reaction with no input DNA. See also Table S2.
Figure 7. HIF1α Activity is Impaired by Copy Number Changes and Mutations in pVHL-Defective Kidney Cancers
(A) Heat map depicting genes that are differentially expressed between VHL−/− renal carcinoma cell lines that do (H1H2) or do not (H2) express high levels of HIF1α. Genes (right) are ordered from top to bottom according to p values showing the degree to which they are significantly altered between H1H2 cells compared to H2 cells. T-test and Hierarchical Clustering was performed using software GENE-E at Broad Institute (57). (B) A Gene Set Enrichment Analysis (GSEA) plot showing the location of the Enrichment Score (ES) of 71 HIF1α up-regulated genes in a set of ccRCC tumors with biallelic VHL inactivation. The HIF1α positively regulated genes are significantly enriched (p ≤ 0.01) in tumors that have not sustained 14q deletions encompassing HIF1α. (C) Schematic of HIF1α mutations identified in renal carcinoma patients. (D and E) Immunoblot (D) and proliferation (E) assays of 769P cells infected with doxycycline-inducible retroviruses encoding wild-type HIF1α or the indicated HIF1α variants and propagated in the presence of doxycyline. Cells grown in the absence of doxycycline were included as controls in (D).
Comment in
- Unraveling the role of hypoxia-inducible factor in renal cell carcinoma: a biological and therapeutic perspective.
Pal SK, Figlin RA. Pal SK, et al. Cancer Discov. 2011 Aug;1(3):198-9. doi: 10.1158/2159-8290.CD-11-0149. Cancer Discov. 2011. PMID: 22586568
Similar articles
- Deletions of chromosomes 3p and 14q molecularly subclassify clear cell renal cell carcinoma.
Kroeger N, Klatte T, Chamie K, Rao PN, Birkhäuser FD, Sonn GA, Riss J, Kabbinavar FF, Belldegrun AS, Pantuck AJ. Kroeger N, et al. Cancer. 2013 Apr 15;119(8):1547-54. doi: 10.1002/cncr.27947. Epub 2013 Jan 18. Cancer. 2013. PMID: 23335244 - HIF1α is not a target of 14q deletion in clear cell renal cancer.
Shenoy N. Shenoy N. Sci Rep. 2020 Oct 19;10(1):17642. doi: 10.1038/s41598-020-74631-7. Sci Rep. 2020. PMID: 33077781 Free PMC article. - Mutant versions of von Hippel-Lindau (VHL) can protect HIF1α from SART1-mediated degradation in clear-cell renal cell carcinoma.
Ordóñez-Navadijo Á, Fuertes-Yebra E, Acosta-Iborra B, Balsa E, Elorza A, Aragonés J, Landazuri MO. Ordóñez-Navadijo Á, et al. Oncogene. 2016 Feb 4;35(5):587-94. doi: 10.1038/onc.2015.113. Epub 2015 Apr 27. Oncogene. 2016. PMID: 25915846 - The role of HIF1α in renal cell carcinoma tumorigenesis.
Gudas LJ, Fu L, Minton DR, Mongan NP, Nanus DM. Gudas LJ, et al. J Mol Med (Berl). 2014 Aug;92(8):825-36. doi: 10.1007/s00109-014-1180-z. Epub 2014 Jun 12. J Mol Med (Berl). 2014. PMID: 24916472 Free PMC article. Review. - The von Hippel-Lindau tumor suppressor gene and kidney cancer.
Kaelin WG Jr. Kaelin WG Jr. Clin Cancer Res. 2004 Sep 15;10(18 Pt 2):6290S-5S. doi: 10.1158/1078-0432.CCR-sup-040025. Clin Cancer Res. 2004. PMID: 15448019 Review.
Cited by
- Research progress on the correlation between obesity and the occurrence and development of kidney cancer: a narrative review.
Kang L, Chen X, Qi P, Ma Z, Han D, Zhang X, Shang P. Kang L, et al. Transl Cancer Res. 2024 Oct 31;13(10):5678-5690. doi: 10.21037/tcr-24-744. Epub 2024 Oct 29. Transl Cancer Res. 2024. PMID: 39525017 Free PMC article. Review. - Toward a CRISPR-based mouse model of _Vhl_-deficient clear cell kidney cancer: Initial experience and lessons learned.
Stransky LA, Gao W, Schmidt LS, Bi K, Ricketts CJ, Ramesh V, James A, Difilippantonio S, Ileva L, Kalen JD, Karim B, Jeon A, Morgan T, Warner AC, Turan S, Unite J, Tran B, Choudhari S, Zhao Y, Linn DE, Yun C, Dhandapani S, Parab V, Pinheiro EM, Morris N, He L, Vigeant SM, Pignon JC, Sticco-Ivins M, Signoretti S, Van Allen EM, Linehan WM, Kaelin WG Jr. Stransky LA, et al. Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2408549121. doi: 10.1073/pnas.2408549121. Epub 2024 Oct 4. Proc Natl Acad Sci U S A. 2024. PMID: 39365820 Free PMC article. - Network modeling links kidney developmental programs and the cancer type-specificity of VHL mutations.
Dong X, Zhang D, Zhang X, Liu Y, Liu Y. Dong X, et al. NPJ Syst Biol Appl. 2024 Oct 3;10(1):114. doi: 10.1038/s41540-024-00445-2. NPJ Syst Biol Appl. 2024. PMID: 39362887 Free PMC article. - Differential effects of hypoxia on motility using various in vitro models of lung adenocarcinoma.
Surguta SE, Baranyi M, Svajda L, Cserepes M, Ranđelović I, Tátrai E, Hegedűs B, Tóvári J. Surguta SE, et al. Sci Rep. 2024 Sep 3;14(1):20482. doi: 10.1038/s41598-024-70769-w. Sci Rep. 2024. PMID: 39227650 Free PMC article. - TFE3 fusions direct an oncogenic transcriptional program that drives OXPHOS and unveils vulnerabilities in translocation renal cell carcinoma.
Li J, Huang K, McBride F, Sadagopan A, Gallant DS, Thakur M, Khanna P, Li B, Ge M, Weiss CN, Achom M, Xu Q, Huang K, Ryback BA, Gui M, Bar-Peled L, Viswanathan SR. Li J, et al. bioRxiv [Preprint]. 2024 Aug 10:2024.08.09.607311. doi: 10.1101/2024.08.09.607311. bioRxiv. 2024. PMID: 39149323 Free PMC article. Preprint.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. - PubMed
- Kim WY, Kaelin WG. Role of VHL Gene Mutation in Human Cancer. J Clin Onc. 2004;22:4991–5004. - PubMed
- Kaelin WG. von Hippel-Lindau Disease. Annual Review of Pathology: Mechanisms of Disease. 2007;2:145–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA068490-15/CA/NCI NIH HHS/United States
- R01 CA068490/CA/NCI NIH HHS/United States
- R01 CA068490-16/CA/NCI NIH HHS/United States
- R01 CA068490-17/CA/NCI NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical