Autophagy proteins regulate the secretory component of osteoclastic bone resorption - PubMed (original) (raw)
Autophagy proteins regulate the secretory component of osteoclastic bone resorption
Carl J DeSelm et al. Dev Cell. 2011.
Erratum in
- Dev Cell. 2011 Dec 13;21(6):1179
Abstract
Osteoclasts resorb bone via the ruffled border, whose complex folds are generated by secretory lysosome fusion with bone-apposed plasma membrane. Lysosomal fusion with the plasmalemma results in acidification of the resorptive microenvironment and release of CatK to digest the organic matrix of bone. The means by which secretory lysosomes are directed to fuse with the ruffled border are enigmatic. We show that proteins essential for autophagy, including Atg5, Atg7, Atg4B, and LC3, are important for generating the osteoclast ruffled border, the secretory function of osteoclasts, and bone resorption in vitro and in vivo. Further, Rab7, which is required for osteoclast function, localizes to the ruffled border in an Atg5-dependent manner. Thus, autophagy proteins participate in polarized secretion of lysosomal contents into the extracellular space by directing lysosomes to fuse with the plasma membrane. These findings are in keeping with a putative link between autophagy genes and human skeletal homeostasis.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures
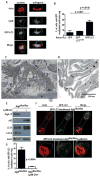
Figure 1. GFP-LC3 localization to the ruffled border is Atg5-dependent
(A) Confocal image of GFP-LC3 transgenic bone-residing osteoclast; CatK, blue; actin, red; GFP, green; cell perimeters, white. CatK and GFP-LC3 localize within the actin ring in the xy and orthogonal planes, representative of two experiments. (B) Percent of osteoclasts with GFP within the actin ring. Osteoclast precursors were transduced with GFP, GFP-LC3 or GFP-LC3G120A and differentiated on bone for 6 days; n=40, pooled from two experiments. (C) Electron microscopy of a resorbing osteoclast on bone (B) with a developed ruffled border (RB) surrounded by actin rings (AR). (D) Immuno-gold labeling of GFP-LC3 in the ruffled border of a resorbing osteoclast. (E) Western blot of Atg5, p62, LC3, and β-actin in control and Atg5-deficient osteoclasts, representative of 6 experiments. (F) Bone-residing control and Atg5-deficient osteoclasts transduced with GFP-LC3 visualized by confocal microscopy; actin, green; CatK, red; cell perimeter, white. (G) Percentage of control and Atg5-deficient osteoclasts with GFP-LC3 concentrated within the actin ring; n=40, pooled from two experiments.
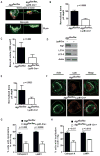
Figure 2. _Atg5_- and _Atg7_-deficient osteoclasts exhibit defective bone pit excavation and lysosome localization to the resorptive microenvironment
(A) Control and Atg5-deficient bone-residing osteoclasts immunostained for actin (red) and exposed to FITC-wheat germ agglutinin (WGA) which stains cells and bone pits green; white, cell perimeters. Bone was visualized in z plane by confocal microscopy before (top) and following removal of cells (bottom), representative of two experiments. (B) Normalized bone pit depth excavated by control and Atg5-deficient osteoclasts. Data from of four experiments (n=100) was normalized to the average depth excavated by control cells as 100% and analyzed using paired t-test. (C) 3D confocal images of FITC-WGA-stained bone pits excavated by control and Atg5-deficient osteoclasts used to determine pit volume; LSM software, one of two experiments shown, n=7 per genotype per experiment. (D) Expression of Atg7, LC3, and β-actin in lysates of control or Atg7-deficient osteoclasts generated for 6 days on plastic; representative of three experiments. (E) Normalized bone pit depth excavated by control and Atg7-deficient osteoclasts; one of two experiments shown, n=7 per genotype per experiment. (F) Representative confocal images of control and Atg5-deficient bone-residing osteoclasts; actin, green; CatK, red; cell perimeters, white. (G) Percentage of control or Atg5-deficient bone-residing osteoclasts with CatK or LAMP1 concentrated within the actin ring; n=45, pooled from three experiments. (H) Percentage of control or Atg7-deficient bone-residing osteoclasts with CatK or LAMP1 localized in the actin ring; n=30, pooled from two experiments.
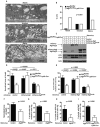
Figure 3. Autophagy proteins mediate ruffled border formation and osteoclast secretory function
(A) Representative electron micrographs of tibia-residing osteoclasts with absent (top), immature (middle), and mature (bottom) ruffled borders adjacent to actin rings (asterisk). (B) Percentage of control or Atg5-deficient osteoclasts exhibiting absent, immature, or mature ruffled borders; representative of two experiments, analyzed by chi-square test for trend, n=30 cells per mouse per genotype. (C) Western blots for Atg5, LC3, or β-actin in control or Atg5-deficient osteoclasts transduced with mCherry-Atg5 or mCherry-Atg5K130R encoding retrovirueses; *, unknown bands, representative of three experiments. (D) Normalized bone pit depth excavated by retrovirally-transduced control or Atg5-deficient osteoclasts; n=20, pooled from three experiments, analyzed by paired t-test. (E) Percentage of osteoclasts with CatK concentrated within the actin ring in retrovirally-transduced control or Atg5-deficient osteoclasts; n=45, pooled from three experiments. (F) Normalized bone pit depth excavated by wildtype osteoclasts retrovirally-transduced with empty vector (control) or mStraberry-Atg4BC74A, expressed as a percentage relative to control; n=30, pooled from three experiments, analyzed by paired t-test. (G) Percentage of cells with CatK localization in actin rings of wildtype osteoclasts transduced with empty vector (control) or Atg4BC74A; n=45, representative of three experiments shown. (H) Percentage of cells with GFP-LC3 localized in the actin ring of vector (control) or Atg4BC74A transduced GFP-LC3 osteoclasts; n=45, representative of two experiments. (I) Percentage of cells with Rab7 localized in the actin ring of control or Atg5-deficient osteoclasts; n=60, pooled from four experiments.
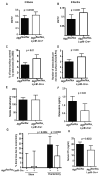
Figure 4. Requirements for Atg5-dependent osteoclast function in vivo
(A, B) Percentage of marrow space occupied by trabecular bone (BV/TV) of 8-week and 8-month old female Atg5flox/flox or _Atg5flox/flox_-LyzM-_Cre_+ mice by microcomputed tomography; n=12 eight week old mice/genotype, n=6 eight-month old Atg5flox/flox mice, n=5 eight month old _Atg5flox/flox_-LyzM-_Cre_+ mice. (C) Percentage of trabecular bone surface covered by TRAP+ osteoclasts determined by histomorphometry; n=5 mice/genotype. (D) Number of osteoclasts per mm of femoral trabecular bone surface determined by histomorphometry; n=5 mice/genotype. (E) Serum osteocalcin levels in 8-week old Atg5flox/flox (n= 5) and _Atg5flox/flox_-LyzM-_Cre_+ mice (n=6) mice. (F) Bone nodule formation by osteoblasts in vitro. Nodules were stained with Alizarin Red-S and quantified using densitometry; pooled from three experiments, n=7 Atg5flox/flox mice, n=10 _Atg5flox/flox_-LyzM-_Cre_+ mice. (G) Percentage of bone loss determined by BV/TV in the four weeks following ovariectomy. Ovariectomy (ovx) or sham operation was performed on Atg5flox/flox (8 ovx, 5 sham) or _Atg5flox/flox_-LyzM-_Cre_+ (7 ovx, 4 sham) mice. The average bone loss of sham-operated Atg5flox/flox animals was subtracted from all measurements. (H) Serum concentration of c-telopeptide of type I collagen (CTx) determined two weeks post-ovariectomy; n=6 mice per genotype.
Comment in
- Autophagic factors cut to the bone.
Gelman A, Elazar Z. Gelman A, et al. Dev Cell. 2011 Nov 15;21(5):808-10. doi: 10.1016/j.devcel.2011.10.021. Dev Cell. 2011. PMID: 22075143
Similar articles
- Aesculetin Inhibits Osteoclastic Bone Resorption through Blocking Ruffled Border Formation and Lysosomal Trafficking.
Na W, Lee EJ, Kang MK, Kim YH, Kim DY, Oh H, Kim SI, Oh SY, Kang YH. Na W, et al. Int J Mol Sci. 2020 Nov 13;21(22):8581. doi: 10.3390/ijms21228581. Int J Mol Sci. 2020. PMID: 33203061 Free PMC article. - Actin-binding protein coronin 1A controls osteoclastic bone resorption by regulating lysosomal secretion of cathepsin K.
Ohmae S, Noma N, Toyomoto M, Shinohara M, Takeiri M, Fuji H, Takemoto K, Iwaisako K, Fujita T, Takeda N, Kawatani M, Aoyama M, Hagiwara M, Ishihama Y, Asagiri M. Ohmae S, et al. Sci Rep. 2017 Mar 16;7:41710. doi: 10.1038/srep41710. Sci Rep. 2017. PMID: 28300073 Free PMC article. - Rab GTPases in Osteoclastic Bone Resorption and Autophagy.
Roy M, Roux S. Roy M, et al. Int J Mol Sci. 2020 Oct 16;21(20):7655. doi: 10.3390/ijms21207655. Int J Mol Sci. 2020. PMID: 33081155 Free PMC article. Review. - Disruption of the dynein-dynactin complex unveils motor-specific functions in osteoclast formation and bone resorption.
Ng PY, Cheng TS, Zhao H, Ye S, Sm Ang E, Khor EC, Feng HT, Xu J, Zheng MH, Pavlos NJ. Ng PY, et al. J Bone Miner Res. 2013 Jan;28(1):119-34. doi: 10.1002/jbmr.1725. J Bone Miner Res. 2013. PMID: 22887640 - The osteoclast and its unique cytoskeleton.
Teitelbaum SL. Teitelbaum SL. Ann N Y Acad Sci. 2011 Dec;1240:14-7. doi: 10.1111/j.1749-6632.2011.06283.x. Ann N Y Acad Sci. 2011. PMID: 22172034 Review.
Cited by
- The functional and pathologic relevance of autophagy proteases.
Fernández ÁF, López-Otín C. Fernández ÁF, et al. J Clin Invest. 2015 Jan;125(1):33-41. doi: 10.1172/JCI73940. Epub 2015 Jan 2. J Clin Invest. 2015. PMID: 25654548 Free PMC article. Review. - The role of kaempferol-induced autophagy on differentiation and mineralization of osteoblastic MC3T3-E1 cells.
Kim IR, Kim SE, Baek HS, Kim BJ, Kim CH, Chung IK, Park BS, Shin SH. Kim IR, et al. BMC Complement Altern Med. 2016 Aug 31;16(1):333. doi: 10.1186/s12906-016-1320-9. BMC Complement Altern Med. 2016. PMID: 27581091 Free PMC article. - Of LAP, CUPS, and DRibbles - Unconventional Use of Autophagy Proteins for MHC Restricted Antigen Presentation.
Münz C. Münz C. Front Immunol. 2015 Apr 29;6:200. doi: 10.3389/fimmu.2015.00200. eCollection 2015. Front Immunol. 2015. PMID: 25972871 Free PMC article. Review. - The Role of TAK1 in RANKL-Induced Osteoclastogenesis.
Jianwei W, Ye T, Hongwei W, Dachuan L, Fei Z, Jianyuan J, Hongli W. Jianwei W, et al. Calcif Tissue Int. 2022 Jul;111(1):1-12. doi: 10.1007/s00223-022-00967-z. Epub 2022 Mar 14. Calcif Tissue Int. 2022. PMID: 35286417 Review. - [Role of autophagy in the pathogenesis of periodontitis].
Mo LY, Jia XY, Liu CC, Zhou XD, Xu X. Mo LY, et al. Hua Xi Kou Qiang Yi Xue Za Zhi. 2019 Aug 1;37(4):422-427. doi: 10.7518/hxkq.2019.04.016. Hua Xi Kou Qiang Yi Xue Za Zhi. 2019. PMID: 31512838 Free PMC article. Review. Chinese.
References
- Cann GM, Guignabert C, Ying L, Deshpande N, Bekker JM, Wang L, Zhou B, Rabinovitch M. Developmental expression of LC3alpha and beta: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev Dyn. 2008;237:187–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI084887/AI/NIAID NIH HHS/United States
- AR032788/AR/NIAMS NIH HHS/United States
- R01 AR046523/AR/NIAMS NIH HHS/United States
- R37 AR046523/AR/NIAMS NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- R01 AI0848887/AI/NIAID NIH HHS/United States
- AR054618/AR/NIAMS NIH HHS/United States
- AR057235/AR/NIAMS NIH HHS/United States
- AR057037/AR/NIAMS NIH HHS/United States
- R01 AR032788/AR/NIAMS NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- P30 AR057235/AR/NIAMS NIH HHS/United States
- R01 AR054618/AR/NIAMS NIH HHS/United States
- F30 AG032795/AG/NIA NIH HHS/United States
- S10 RR023660/RR/NCRR NIH HHS/United States
- AR046523/AR/NIAMS NIH HHS/United States
- AG032795/AG/NIA NIH HHS/United States
- R01 AR057037/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases