Vanillin catabolism in Rhodococcus jostii RHA1 - PubMed (original) (raw)
Vanillin catabolism in Rhodococcus jostii RHA1
Hao-Ping Chen et al. Appl Environ Microbiol. 2012 Jan.
Abstract
Genes encoding vanillin dehydrogenase (vdh) and vanillate O-demethylase (vanAB) were identified in Rhodococcus jostii RHA1 using gene disruption and enzyme activities. During growth on vanillin or vanillate, vanA was highly upregulated while vdh was not. This study contributes to our understanding of lignin degradation by RHA1 and other actinomycetes.
Figures
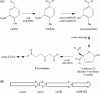
Fig 1
(A) Vanillin catabolic pathway in RHA1. (B) Vanillate _O_-demethylase gene cluster. The vanA and vanB genes encode the oxygenase and reductase components, respectively, of vanillate _O_-demethylase. Gene ro04162 is predicted to encode an IclR family transcriptional regulator. Acetyl-CoA, acetyl coenzyme A.
Similar articles
- The catabolism of lignin-derived _p_-methoxylated aromatic compounds by Rhodococcus jostii RHA1.
Wolf ME, Lalande AT, Newman BL, Bleem AC, Palumbo CT, Beckham GT, Eltis LD. Wolf ME, et al. Appl Environ Microbiol. 2024 Mar 20;90(3):e0215523. doi: 10.1128/aem.02155-23. Epub 2024 Feb 21. Appl Environ Microbiol. 2024. PMID: 38380926 Free PMC article. - Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate.
Priefert H, Rabenhorst J, Steinbüchel A. Priefert H, et al. J Bacteriol. 1997 Apr;179(8):2595-607. doi: 10.1128/jb.179.8.2595-2607.1997. J Bacteriol. 1997. PMID: 9098058 Free PMC article. - Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin.
Spence EM, Calvo-Bado L, Mines P, Bugg TDH. Spence EM, et al. Microb Cell Fact. 2021 Jan 19;20(1):15. doi: 10.1186/s12934-020-01504-z. Microb Cell Fact. 2021. PMID: 33468127 Free PMC article. - The Hydroxyquinol Degradation Pathway in Rhodococcus jostii RHA1 and Agrobacterium Species Is an Alternative Pathway for Degradation of Protocatechuic Acid and Lignin Fragments.
Spence EM, Scott HT, Dumond L, Calvo-Bado L, di Monaco S, Williamson JJ, Persinoti GF, Squina FM, Bugg TDH. Spence EM, et al. Appl Environ Microbiol. 2020 Sep 17;86(19):e01561-20. doi: 10.1128/AEM.01561-20. Print 2020 Sep 17. Appl Environ Microbiol. 2020. PMID: 32737130 Free PMC article. - Adventures in Rhodococcus - from steroids to explosives.
Yam KC, Okamoto S, Roberts JN, Eltis LD. Yam KC, et al. Can J Microbiol. 2011 Mar;57(3):155-68. doi: 10.1139/W10-115. Can J Microbiol. 2011. PMID: 21358756 Review.
Cited by
- The catabolism of lignin-derived _p_-methoxylated aromatic compounds by Rhodococcus jostii RHA1.
Wolf ME, Lalande AT, Newman BL, Bleem AC, Palumbo CT, Beckham GT, Eltis LD. Wolf ME, et al. Appl Environ Microbiol. 2024 Mar 20;90(3):e0215523. doi: 10.1128/aem.02155-23. Epub 2024 Feb 21. Appl Environ Microbiol. 2024. PMID: 38380926 Free PMC article. - Harnessing redox proteomics to study metabolic regulation and stress response in lignin-fed Rhodococci.
Li X, Gluth A, Feng S, Qian WJ, Yang B. Li X, et al. Biotechnol Biofuels Bioprod. 2023 Nov 20;16(1):180. doi: 10.1186/s13068-023-02424-x. Biotechnol Biofuels Bioprod. 2023. PMID: 37986172 Free PMC article. - A key _O_-demethylase in the degradation of guaiacol by Rhodococcus opacus PD630.
Xue L, Zhao Y, Li L, Rao X, Chen X, Ma F, Yu H, Xie S. Xue L, et al. Appl Environ Microbiol. 2023 Oct 31;89(10):e0052223. doi: 10.1128/aem.00522-23. Epub 2023 Oct 6. Appl Environ Microbiol. 2023. PMID: 37800939 Free PMC article. - Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on _Meta_-Methoxylated Aromatic Acids.
Donoso RA, Corbinaud R, Gárate-Castro C, Galaz S, Pérez-Pantoja D. Donoso RA, et al. Microorganisms. 2022 Dec 27;11(1):78. doi: 10.3390/microorganisms11010078. Microorganisms. 2022. PMID: 36677370 Free PMC article. - Bacterial catabolism of acetovanillone, a lignin-derived compound.
Dexter GN, Navas LE, Grigg JC, Bajwa H, Levy-Booth DJ, Liu J, Louie NA, Nasseri SA, Jang SK, Renneckar S, Eltis LD, Mohn WW. Dexter GN, et al. Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2213450119. doi: 10.1073/pnas.2213450119. Epub 2022 Oct 18. Proc Natl Acad Sci U S A. 2022. PMID: 36256818 Free PMC article.
References
- Ahmad M, et al. 2011. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–5107 - PubMed
- Ahmad M, et al. 2010. Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders, Mol. Biosyst. 6:815–821 - PubMed
- Bauchop T, Elsden SR. 1960. The growth of micro-organisms in relation to their energy supply. J. Gen. Microbiol. 23:457–469 - PubMed
- Bugg TD, Ahmad M, Hardiman EM, Singh R. 2011. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22:394–400 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources