Malaria antifolate resistance with contrasting Plasmodium falciparum dihydrofolate reductase (DHFR) polymorphisms in humans and Anopheles mosquitoes - PubMed (original) (raw)
Malaria antifolate resistance with contrasting Plasmodium falciparum dihydrofolate reductase (DHFR) polymorphisms in humans and Anopheles mosquitoes
Sungano Mharakurwa et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Surveillance for drug-resistant parasites in human blood is a major effort in malaria control. Here we report contrasting antifolate resistance polymorphisms in Plasmodium falciparum when parasites in human blood were compared with parasites in Anopheles vector mosquitoes from sleeping huts in rural Zambia. DNA encoding P. falciparum dihydrofolate reductase (EC 1.5.1.3) was amplified by PCR with allele-specific restriction enzyme digestions. Markedly prevalent pyrimethamine-resistant mutants were evident in human P. falciparum infections--S108N (>90%), with N51I, C59R, and 108N+51I+59R triple mutants (30-80%). This resistance level may be from selection pressure due to decades of sulfadoxine/pyrimethamine use in the region. In contrast, cycloguanil-resistant mutants were detected in very low frequency in parasites from human blood samples-S108T (13%), with A16V and 108T+16V double mutants (∼4%). Surprisingly, pyrimethamine-resistant mutants were of very low prevalence (2-12%) in the midguts of Anopheles arabiensis vector mosquitoes, but cycloguanil-resistant mutants were highly prevalent-S108T (90%), with A16V and the 108T+16V double mutant (49-57%). Structural analysis of the dihydrofolate reductase by in silico modeling revealed a key difference in the enzyme within the NADPH binding pocket, predicting the S108N enzyme to have reduced stability but the S108T enzyme to have increased stability. We conclude that P. falciparum can bear highly host-specific drug-resistant polymorphisms, most likely reflecting different selective pressures found in humans and mosquitoes. Thus, it may be useful to sample both human and mosquito vector infections to accurately ascertain the epidemiological status of drug-resistant alleles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
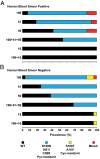
Fig. 1.
Prevalence of P. falciparum pyrimethamine- and cycloguanil-resistant DHFR mutants in human blood. Finger-prick samples were obtained from humans with microscopic parasitemia (blood film-positive; n = 86; A) and submicroscopic parasitemia (blood film-negative; n = 88; B). DNA was PCR amplified and analyzed for allele-specific polymorphisms by restriction digestion. _y_-axis numerals denote DHFR codons. Represented are WT, pyrimethamine-resistant mutants (S108N, N51I, C59R, and triple mutants), cycloguanil-resistant mutants (S108T, A16V, and double mutant), and mixed alleles (WT+S108T or S108N+S108T).
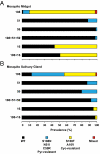
Fig. 2.
Prevalence of P. falciparum pyrimethamine- and cycloguanil-resistant DHFR mutants in 796 malaria vector mosquitoes. Spray catches were obtained. Midguts (A) and salivary gland (B) specimens were separated, and DNA was PCR amplified and analyzed for allele-specific polymorphisms by restriction digestion confirming midgut infections (n = 81) and mosquito gland infections (n = 62). _y_-axis numerals denote DHFR codons. Represented are WT, pyrimethamine-resistant mutants (S108N, N51I, C59R, and triple mutants), cycloguanil-resistant mutants (S108T, A16V, and double mutant), and mixed alleles (WT+S108T or designated mutant+S108T).
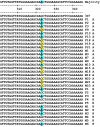
Fig. 3.
DNA sequence alignments for P. falciparum DHFR. DNA flanking nucleotide 323 (amino acid codon 108) obtained from amplicons M3-F/ (M) and F-M4 (F) from human (H) and Anopheles mosquito (A) samples are plotted against standard clone 3D7 (GenBank accession no. AL844503; gene ID 812524). Nucleotide 323 encoding amino acid 108 is accentuated. AGC encodes WT S108; AAC encodes Pyr-resistant mutant S108N (blue); ACC encodes Cyc-resistant mutant S108T (yellow). Identical DNA sequences were obtained from both duplicate amplicons except one mosquito set (F19 A, M19 A) and one human set (F2 H, M2 H), which contained mixed infections. All results confirmed the PCR and restriction enzyme typing.
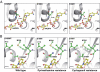
Fig. 4.
Key structural variances at amino acid 108 in WT and resistant mutants of P. falciparum DHFR. Represented are crystal structures of WT DHFR (S108WT) and S108N mutant (PDB ID codes 1J3I and 3JSU). The structure of the S108T mutant was derived by molecular modeling. (A) Note a 2.56-Å hydrogen bond between the pyrophosphate moiety of NADPH and the hydroxyl group of the S108 WT or S108T residue; no effective hydrogen bond exists in the S108N mutant due to the 5.40-Å distance. (B) Note extra methyl group in the S108T mutant forms van der Waals interactions with a nearby hydrophobic area composed of Met-104, Ile-112, and Ile-164. The side chains of S108WT, S108T, S108N, Met-104, Ile-112, Ile-164, and NADPH are highlighted with sticks and spheres; the hydrogen bond is shown as the dashed line with the distance labeled, and the protein backbone is shown in a cartoon model.
Similar articles
- Novel alleles of Plasmodium falciparum dhfr that confer resistance to chlorcycloguanil.
Hunt SY, Rezvani BB, Sibley CH. Hunt SY, et al. Mol Biochem Parasitol. 2005 Jan;139(1):25-32. doi: 10.1016/j.molbiopara.2004.09.005. Mol Biochem Parasitol. 2005. PMID: 15610816 - Pyrimethamine analogs as strong inhibitors of double and quadruple mutants of dihydrofolate reductase in human malaria parasites.
Sardarian A, Douglas KT, Read M, Sims PF, Hyde JE, Chitnumsub P, Sirawaraporn R, Sirawaraporn W. Sardarian A, et al. Org Biomol Chem. 2003 Mar 21;1(6):960-4. doi: 10.1039/b211636g. Org Biomol Chem. 2003. PMID: 12929634 - The dihydrofolate reductase-thymidylate synthetase gene in the drug resistance of malaria parasites.
Hyde JE. Hyde JE. Pharmacol Ther. 1990;48(1):45-59. doi: 10.1016/0163-7258(90)90017-v. Pharmacol Ther. 1990. PMID: 2274577 Review.
Cited by
- The molecular basis of antifolate resistance in Plasmodium falciparum: looking beyond point mutations.
Heinberg A, Kirkman L. Heinberg A, et al. Ann N Y Acad Sci. 2015 Apr;1342(1):10-8. doi: 10.1111/nyas.12662. Epub 2015 Feb 18. Ann N Y Acad Sci. 2015. PMID: 25694157 Free PMC article. Review. - Transmission of molecularly undetectable circulating parasite clones leads to high infection complexity in mosquitoes post feeding.
Grignard L, Gonçalves BP, Early AM, Daniels RF, Tiono AB, Guelbéogo WM, Ouédraogo A, van Veen EM, Lanke K, Diarra A, Nebie I, Sirima SB, Targett GA, Volkman SK, Neafsey DE, Wirth DF, Bousema T, Drakeley C. Grignard L, et al. Int J Parasitol. 2018 Jul;48(8):671-677. doi: 10.1016/j.ijpara.2018.02.005. Epub 2018 May 5. Int J Parasitol. 2018. PMID: 29738740 Free PMC article. - Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae.
Gómez-Díaz E, Yerbanga RS, Lefèvre T, Cohuet A, Rowley MJ, Ouedraogo JB, Corces VG. Gómez-Díaz E, et al. Sci Rep. 2017 Jan 16;7:40655. doi: 10.1038/srep40655. Sci Rep. 2017. PMID: 28091569 Free PMC article. - Modelling the impact of antimalarial quality on the transmission of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum.
Brock AR, Ross JV, Greenhalgh S, Durham DP, Galvani A, Parikh S, Esterman A. Brock AR, et al. Infect Dis Model. 2017 Apr 15;2(2):161-187. doi: 10.1016/j.idm.2017.04.001. eCollection 2017 May. Infect Dis Model. 2017. PMID: 29928735 Free PMC article. - The Rare, the Best: Spread of Antimalarial-Resistant Plasmodium falciparum Parasites by Anopheles Mosquito Vectors.
Berry A, Menard S, Nsango SE, Abate L, Concordet D, Tchioffo Tsapi M, Iriart X, Awono-Ambéné PH, Roche B, Morlais I. Berry A, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0085221. doi: 10.1128/Spectrum.00852-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668767 Free PMC article.
References
- Björkman A, Bhattarai A. Public health impact of drug resistant Plasmodium falciparum malaria. Acta Trop. 2005;94:163–169. - PubMed
- Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64(1–2 Suppl):12–17. - PubMed
- Noedl H, et al. Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical