Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems - PubMed (original) (raw)
Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems
André E Minoche et al. Genome Biol. 2011.
Abstract
Background: The generation and analysis of high-throughput sequencing data are becoming a major component of many studies in molecular biology and medical research. Illumina's Genome Analyzer (GA) and HiSeq instruments are currently the most widely used sequencing devices. Here, we comprehensively evaluate properties of genomic HiSeq and GAIIx data derived from two plant genomes and one virus, with read lengths of 95 to 150 bases.
Results: We provide quantifications and evidence for GC bias, error rates, error sequence context, effects of quality filtering, and the reliability of quality values. By combining different filtering criteria we reduced error rates 7-fold at the expense of discarding 12.5% of alignable bases. While overall error rates are low in HiSeq data we observed regions of accumulated wrong base calls. Only 3% of all error positions accounted for 24.7% of all substitution errors. Analyzing the forward and reverse strands separately revealed error rates of up to 18.7%. Insertions and deletions occurred at very low rates on average but increased to up to 2% in homopolymers. A positive correlation between read coverage and GC content was found depending on the GC content range.
Conclusions: The errors and biases we report have implications for the use and the interpretation of Illumina sequencing data. GAIIx and HiSeq data sets show slightly different error profiles. Quality filtering is essential to minimize downstream analysis artifacts. Supporting previous recommendations, the strand-specificity provides a criterion to distinguish sequencing errors from low abundance polymorphisms.
Figures
Figure 1
Distribution of read coverage depth for (a) Bv-95nt reads and (b) Phix-95nt reads. Read coverage was computed per base. In three separate calculations we considered all positions (black), positions in regions below (red) and positions in regions above (blue) the average GC content (%GC) of the reference. The regional %GC was determined based on a window of 250 bases upstream and 250 bases downstream of each position. In contrast to PhiX (b) the coverage variation in the sugar beet sample (a) is related to the %GC.
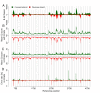
Figure 2
Distribution of low quality bases along the PhiX reference genome. Analysis was performed on reads derived from an Illumina PhiX library (PhiX-95nt data set). (a) Number of bases within B-tails (consecutive bases of Q-score = 2 at the 3' end of a read) per position. (b) Average Q-score of bases in untrimmed reads. (c) Average Q-score of bases in B-tail trimmed reads. (d) Observed per-base substitution error rate. Calculations for (a-d) were performed separately for the forward strand (green) and reverse strand (red). Low quality values accumulated in certain regions even after removal of B-tails. The peaks of observed error rates occur at positions where increased low quality counts are detected, and in most cases the peak is seen only on one strand.
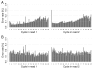
Figure 3
Alignment of reads before (a) and after (b) B-tail trimming in a selected region of the ZR reference (positions 63,633 to 63,662). Uniquely mapped reads from the Bv-95nt data set were visualized using the Tablet browser [17]. Forward matching reads are shown in grey, reverse matching reads are shown in blue, mismatch bases are shown in white. Long white stretches are uncalled bases. Mismatches accumulated in one region, and almost all mismatches are eliminated after B-tail removal.
Figure 4
Observed error rates of 2 × 95-nucleotide HiSeq reads by cycle (averaged across all flow cell tiles). Read 1 (left) and read 2 (right) were analyzed separately for PhiX-95nt data (a) and Bv-95nt data (b). PhiX and sugar beet DNA was sequenced in the same lane, and reads were mapped against the PhiX or ZR reference sequences, respectively.
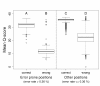
Figure 5
Mean quality scores of correct and wrong bases sequenced (PhiX-95nt data) at error-prone positions and all other positions in the PhiX reference. The bases covering 161 positions of significantly elevated error rates (A, B) in the PhiX reference show lower average quality scores compared to the bases covering other positions (C, D). This is true for correctly called bases (A, C) as well as for incorrectly called bases (B, D).
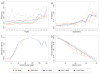
Figure 6
Frequencies and context of sequencing errors and quality scores compared to observed error rates. The sugar beet sample (yellow) and the Arabidopsis sample (blue) were each sequenced together with PhiX DNA (red and green, respectively) on a HiSeq2000 sequencing instrument. PhiX DNA only (black) was sequenced on a GAIIx. (a) Sequence context of substitution errors. The frequency of neighboring bases one position upstream and downstream of an error position is displayed. Sequence triplets were summarized for all types of base substitutions at the central position (indicated by an 'e'). We counted reads spanning the triplet positions and ignored potential further substitution errors within the triplet sequence of the read. The frequency was determined by dividing the occurrence of a triplet containing a central substitution error by the occurrence of all triplets with the same marginal bases but variable central base. The display of triplets is ordered by increasing average frequency in the HiSeq data. (b) Frequency of base substitution errors. For each sample, the proportion of each substitution is indicated (ordered by increasing average frequency in the HiSeq samples). (c) Rates of insertions or deletions in homopolymer tracts normalized by homopolymer length. Homopolymers longer than seven bases were present only in the two plant samples. Homopolymers of length 16 to 19 in the Bv-95nt data and of length 26 to 29 in the At-100nt data were each covered by less than 50 reads. (d) Expected versus observed error rates. Expected error rates according to quality scores (Q) were calculated for Q = 2 to Q = 40 (solid diagonal line). For each sample the uniquely aligned bases were grouped by quality score, and the observed error rate was determined from the number of observed substitution errors for each Q separately.
Similar articles
- Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data.
Schirmer M, D'Amore R, Ijaz UZ, Hall N, Quince C. Schirmer M, et al. BMC Bioinformatics. 2016 Mar 11;17:125. doi: 10.1186/s12859-016-0976-y. BMC Bioinformatics. 2016. PMID: 26968756 Free PMC article. - Substantial biases in ultra-short read data sets from high-throughput DNA sequencing.
Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Dohm JC, et al. Nucleic Acids Res. 2008 Sep;36(16):e105. doi: 10.1093/nar/gkn425. Epub 2008 Jul 26. Nucleic Acids Res. 2008. PMID: 18660515 Free PMC article. - Characterization of sequence-specific errors in various next-generation sequencing systems.
Shin S, Park J. Shin S, et al. Mol Biosyst. 2016 Mar;12(3):914-22. doi: 10.1039/c5mb00750j. Mol Biosyst. 2016. PMID: 26790373 - MiSeq: A Next Generation Sequencing Platform for Genomic Analysis.
Ravi RK, Walton K, Khosroheidari M. Ravi RK, et al. Methods Mol Biol. 2018;1706:223-232. doi: 10.1007/978-1-4939-7471-9_12. Methods Mol Biol. 2018. PMID: 29423801 Review. - Massively parallel sequencing approaches for characterization of structural variation.
Koboldt DC, Larson DE, Chen K, Ding L, Wilson RK. Koboldt DC, et al. Methods Mol Biol. 2012;838:369-84. doi: 10.1007/978-1-61779-507-7_18. Methods Mol Biol. 2012. PMID: 22228022 Free PMC article. Review.
Cited by
- A thesaurus of genetic variation for interrogation of repetitive genomic regions.
Kerzendorfer C, Konopka T, Nijman SM. Kerzendorfer C, et al. Nucleic Acids Res. 2015 May 26;43(10):e68. doi: 10.1093/nar/gkv178. Epub 2015 Mar 27. Nucleic Acids Res. 2015. PMID: 25820428 Free PMC article. - Genome-Centric Dynamics Shape the Diversity of Oral Bacterial Populations.
Utter DR, Cavanaugh CM, Borisy GG. Utter DR, et al. mBio. 2022 Dec 20;13(6):e0241422. doi: 10.1128/mbio.02414-22. Epub 2022 Oct 10. mBio. 2022. PMID: 36214570 Free PMC article. - Molecular Characterization and Subtyping of Breast Cancer Cell Lines Provide Novel Insights into Cancer Relevant Genes.
Pommerenke C, Nagel S, Haake J, Koelz AL, Christgen M, Steenpass L, Eberth S. Pommerenke C, et al. Cells. 2024 Feb 6;13(4):301. doi: 10.3390/cells13040301. Cells. 2024. PMID: 38391914 Free PMC article. - Analysis of the cystic fibrosis lung microbiota via serial Illumina sequencing of bacterial 16S rRNA hypervariable regions.
Maughan H, Wang PW, Diaz Caballero J, Fung P, Gong Y, Donaldson SL, Yuan L, Keshavjee S, Zhang Y, Yau YC, Waters VJ, Tullis DE, Hwang DM, Guttman DS. Maughan H, et al. PLoS One. 2012;7(10):e45791. doi: 10.1371/journal.pone.0045791. Epub 2012 Oct 2. PLoS One. 2012. PMID: 23056217 Free PMC article. - A Deep-Sequencing Workflow for the Fast and Efficient Generation of High-Quality African Swine Fever Virus Whole-Genome Sequences.
Forth JH, Forth LF, King J, Groza O, Hübner A, Olesen AS, Höper D, Dixon LK, Netherton CL, Rasmussen TB, Blome S, Pohlmann A, Beer M. Forth JH, et al. Viruses. 2019 Sep 11;11(9):846. doi: 10.3390/v11090846. Viruses. 2019. PMID: 31514438 Free PMC article.
References
- GenomeWeb. http://www.genomeweb.com/
- Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, Magrini VJ, Richt RJ, Sander SN, Stewart DA, Stromberg M, Tsung EF, Wylie T, Schedl T, Wilson RK, Mardis ER. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous