Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β - PubMed (original) (raw)
Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β
Nicolas Dupont et al. EMBO J. 2011.
Abstract
Autophagy controls the quality and quantity of the eukaryotic cytoplasm while performing two evolutionarily highly conserved functions: cell-autonomous provision of energy and nutrients by cytosol autodigestion during starvation, and removal of defunct organelles and large aggregates exceeding the capacity of other cellular degradative systems. In contrast to these autodigestive processes, autophagy in yeast has additional, biogenesis functions. However, no equivalent biosynthetic roles have been described for autophagy in mammals. Here, we show that in mammalian cells, autophagy has a hitherto unappreciated positive contribution to the biogenesis and secretion of the proinflammatory cytokine IL-1β via an export pathway that depends on Atg5, inflammasome, at least one of the two mammalian Golgi reassembly stacking protein (GRASP) paralogues, GRASP55 (GORASP2) and Rab8a. This process, which is a type of unconventional secretion, expands the functional manifestations of autophagy beyond autodigestive and quality control roles in mammals. It enables a subset of cytosolic proteins devoid of signal peptide sequences, and thus unable to access the conventional pathway through the ER, to enter an autophagy-based secretory pathway facilitating their exit from the cytoplasm.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
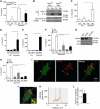
Induction of autophagy enhances IL-1β secretion. (A) Atg5fl/fl Cre− and Atg5fl/fl Cre+ bone marrow-derived macrophages (BMMs), pretreated overnight with 100 ng/ml LPS, were stimulated for 1 h with the inflammasome agonist nigericin (20 μM) with (Starvation; EBSS) or without (Full; full medium) autophagic induction. Cell culture supernatants were assayed for murine IL-1β by ELISA. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (B) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and the release of active caspase-1 and IL-1β was determined by immunoblotting. (C) As in (A), assayed for IL-18. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (D) LPS-pretreated BMMs were exposed to alum (250 μg/ml) for 1 h with or without autophagic induction by starvation. Secreted IL-1β was measured as in (A). Data represent mean values±s.d. (_n_⩾3); *P<0.05. (E) LPS-pretreated BMMs were exposed to silica (250 μg/ml) for 1 h with or without autophagic induction by starvation. Secreted IL-1β was measured as in (A). Data represent mean values±s.d. (_n_⩾3); *P<0.05. (F) BMMs were transfected with scramble (Scr) control siRNA or siRNAs against ASC and NLRP3. After 48 h following transfection, cells were treated overnight with LPS and subjected to nigericin (20 μM) and starvation for 1 h. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (G) Immunoblot analysis of ASC and NLRP3 knockdowns. (H) BMMs were transfected with scramble (Scr) control siRNA or siRNAs against ASC and NLRP3. After 48 h following transfection, cells were treated overnight with LPS and subjected to silica (250 μg/ml) and starvation for 1 h. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (I) Colocalization of IL-1β with the basal autophagic machinery factor LC3. Fluorescence: LC3 (green, Alexa488); IL-1β (red, Alexa568). BMMs were from GFP–LC3 knock-in mice, treated with LPS then prepared for immunofluorescence microscopy using fluorescently labelled antibodies against GFP and IL-1β. (J, K) A line fluorescence tracing from images in (I). (L) Pearson's colocalization coefficient for IL-1β and LC3. Pearson's coefficient was derived from three independent experiments with five fields per experiment, for a total of 15 fields contributing to the cumulative result. Figure source data can be found in Supplementary data.
Figure 2
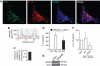
Autophagic pathway progression promotes secretion of inflammasome substrates. (A, B) LPS-pretreated BMMs were treated with 20 μM nigericin (Nig) and 100 nM bafilomycin A1 (Baf) with (Starvation) or without (Full) autophagic induction for 1 h and secreted IL-1β (A) and IL-18 (B) were measured. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (C) LPS-pretreated BMMs were treated with 250 μg/ml of silica and 100 nM bafilomycin A1 (Baf) with (Starvation) or without (Full) autophagic induction for 1 h and secreted IL-1β were measured. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (D) Colocalization of cathepsin B with the basal autophagic machinery factor LC3 and IL-1β. Fluorescence; LC3 (green, Alexa488), IL-1β (red, Alexa568), and cathepsin B (blue, Alexa633). BMMs from GFP–LC3 knock-in mice were treated with LPS and then analysed for immunofluorescence. (E) Colocalization line tracing analysis from images in (D). (F) LPS-pretreated BMMs were treated with 20 μM nigericin and cathepsin B inhibitor CA-074 Me (10 μM), with (Starvation) or without (Full) autophagic induction, for 1 h and secreted IL-1β was measured. Data represent mean values±s.d. (_n_⩾3); *P<0.05. (G) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and release of cathepsin B was determined by immunoblotting. Figure source data can be found in Supplementary data.
Figure 3
Rab8a is required for autophagy-activated IL-1β secretion. (A) Colocalization of Rab8a with the basal autophagic machinery factor LC3 and IL-1β. Fluorescence; LC3 (green, Alexa488), IL-1β (red, Alexa568), Rab8a (blue, Alexa633). BMMs from GFP–LC3 knock-in mice were pretreated with LPS and analysed by immunofluorescence microscopy. Arrows indicate triple colocalization. (B) Line tracing analysis of fluorescence signal intensity. (C) Pearson's colocalization coefficient for IL-1β and Rab8a. Pearson's coefficients were derived from three completely independent experiments with >5 fields per experiment, for a total of ⩾15 fields contributing to the cumulative result. (D) BMMs were transfected with siRNAs against Rab8a or scramble (Scr) control. At 24 h after the first transfection, cells were transfected again with siRNA, treated with LPS and the day after subjected to nigericin in full medium for 1 h, and IL-1β secretion measured. (E) Immunoblot analysis of Rab8a knockdown in BMMs. (F) RAW 264.7 macrophages were transfected with GFP-tagged Rab8a constructs (WT, wild type; S22N, dominant-negative mutant), treated overnight with LPS and stimulated for 1 h with 20 μM nigericin along with induction of autophagy by starvation. IL-1β secretion was measured by ELISA. Data represent mean values±s.d. (_n_⩾3); *P<0.05. Figure source data can be found in Supplementary data.
Figure 4
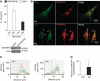
GRASP55 is required for autophagy-activated IL-1β secretion. (A) BMM cells were transfected with scramble (Scr) control siRNA or siRNA against GRASP55. After 48 h of transfection, cells were treated with LPS and the day after subjected to 20 μM nigericin in EBSS, and secreted IL-1β was measured by ELISA. Data represent mean values±s.d. (_n_⩾3); *P<0.05. Inset: Immunoblot analysis of GRASP55 knockdown. (B) Immunofluorescence confocal microscopy analysis of LC3 and GRASP55 distribution. LC3 (green, Alexa488), GRASP55 (red, Alexa568). BMMs were pretreated overnight with 100 ng/ml LPS and either not stimulated (Ctrl) or stimulated (Nig) for 30 min with the inflammasome agonist nigericin (20 μM) in full medium. (C) Line tracings, analysis of fluorescence signal intensity from images in (B). (D) Pearson's coefficients for LC3 and GRASP55 were quantified using SlideBook morphometric analysis software as a measure of adjacency between GRASP55 and LC3 profiles. Pearson's coefficients were derived from three independent experiments with five fields per experiment, for a total of 15 fields contributing to the cumulative result. Figure source data can be found in Supplementary data.
Figure 5
GRASP55 controls autophagy initiation. (A, B) Effect of GRASP55 on autophagy induction by measuring LC3-II. BMM cells were transfected with GRASP55 siRNAs or scramble (Scr) control. At 72 h post transfection, cells were induced for autophagy, treated or not with Bafilomycin A1 (Baf) to inhibit autophagic degradation and LC3-II/actin ratios determined by immunoblotting (A) followed by densitometry (B). Data represent mean values±s.d. (_n_⩾3); *P<0.05. (C, D) RAW 264.7 was transfected with GRASP55 siRNAs or scramble (Scr) siRNA control. Following 48 h of siRNA treatment, cells were transfected with RFP–GFP–LC3 plasmid (GFP is sensitive to acidification, whereas RFP is not), after 24 h induced for autophagy in EBSS for 1 h and autophagic induction and flux quantified (graph in D) by determining the number of early autophagic organelles (GFP+RFP+ puncta) and autolysosomal organelles (GFP−RFP+ puncta) per cell as illustrated in fluorescent images (yellow arrows, GFP+RFP+; red arrows, GFP−RFP+). Total, yellow+red puncta per cell. Data represent mean values±s.d. (_n_⩾3); *P<0.05. Figure source data can be found in Supplementary data.
Figure 6
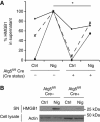
HMGB1 is an autophagy-based alternative secretion substrate. (A) Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs, pretreated overnight with 100 ng/ml LPS, were stimulated for 1 h with 20 μM nigericin (Nig; inflammasome agonist) while incubated in EBSS for induction of autophagy by starvation. Cell culture supernatants were assayed for murine HMGB1 by ELISA. Data (normalized to sample with maximum HMGB1 secretion in each experimental repeat; Cre−and Nig) represent mean values±s.d. (_n_⩾3); *P<0.05. (B) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and the release of HMGB1 was determined by immunoblotting. Figure source data can be found in Supplementary data.
Similar articles
- Autophagy-based unconventional secretion of HMGB1 by keratinocytes plays a pivotal role in psoriatic skin inflammation.
Wang Z, Zhou H, Zheng H, Zhou X, Shen G, Teng X, Liu X, Zhang J, Wei X, Hu Z, Zeng F, Hu Y, Hu J, Wang X, Chen S, Cheng J, Zhang C, Gui Y, Zou S, Hao Y, Zhao Q, Wu W, Zhou Y, Cui K, Huang N, Wei Y, Li W, Li J. Wang Z, et al. Autophagy. 2021 Feb;17(2):529-552. doi: 10.1080/15548627.2020.1725381. Epub 2020 Feb 16. Autophagy. 2021. PMID: 32019420 Free PMC article. - Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators.
Jiang S, Dupont N, Castillo EF, Deretic V. Jiang S, et al. J Innate Immun. 2013;5(5):471-9. doi: 10.1159/000346707. Epub 2013 Feb 22. J Innate Immun. 2013. PMID: 23445716 Free PMC article. Review. - Autophagy Mediates Interleukin-1β Secretion in Human Neutrophils.
Iula L, Keitelman IA, Sabbione F, Fuentes F, Guzman M, Galletti JG, Gerber PP, Ostrowski M, Geffner JR, Jancic CC, Trevani AS. Iula L, et al. Front Immunol. 2018 Feb 19;9:269. doi: 10.3389/fimmu.2018.00269. eCollection 2018. Front Immunol. 2018. PMID: 29515581 Free PMC article. - Secretory autophagy machinery and vesicular trafficking are involved in HMGB1 secretion.
Kim YH, Kwak MS, Lee B, Shin JM, Aum S, Park IH, Lee MG, Shin JS. Kim YH, et al. Autophagy. 2021 Sep;17(9):2345-2362. doi: 10.1080/15548627.2020.1826690. Epub 2020 Oct 5. Autophagy. 2021. PMID: 33017561 Free PMC article. - Evolution, role in inflammation, and redox control of leaderless secretory proteins.
Sitia R, Rubartelli A. Sitia R, et al. J Biol Chem. 2020 May 29;295(22):7799-7811. doi: 10.1074/jbc.REV119.008907. Epub 2020 Apr 24. J Biol Chem. 2020. PMID: 32332096 Free PMC article. Review.
Cited by
- Emerging degrader technologies engaging lysosomal pathways.
Ding Y, Xing D, Fei Y, Lu B. Ding Y, et al. Chem Soc Rev. 2022 Oct 31;51(21):8832-8876. doi: 10.1039/d2cs00624c. Chem Soc Rev. 2022. PMID: 36218065 Free PMC article. Review. - Lysosomal trafficking of TGFBIp via caveolae-mediated endocytosis.
Choi SI, Maeng YS, Kim TI, Lee Y, Kim YS, Kim EK. Choi SI, et al. PLoS One. 2015 Apr 8;10(4):e0119561. doi: 10.1371/journal.pone.0119561. eCollection 2015. PLoS One. 2015. PMID: 25853243 Free PMC article. - Homing of Antigen-Presenting Cells in Head Kidney and Spleen - Salmon Head Kidney Hosts Diverse APC Types.
Iliev DB, Thim H, Lagos L, Olsen R, Jørgensen JB. Iliev DB, et al. Front Immunol. 2013 Jun 6;4:137. doi: 10.3389/fimmu.2013.00137. eCollection 2013. Front Immunol. 2013. PMID: 23761795 Free PMC article. - Autophagy: a new target or an old strategy for the treatment of Crohn's disease?
Nys K, Agostinis P, Vermeire S. Nys K, et al. Nat Rev Gastroenterol Hepatol. 2013 Jul;10(7):395-401. doi: 10.1038/nrgastro.2013.66. Epub 2013 Apr 16. Nat Rev Gastroenterol Hepatol. 2013. PMID: 23591407 Review. - Autophagy and autoimmunity crosstalks.
Bhattacharya A, Eissa NT. Bhattacharya A, et al. Front Immunol. 2013 Apr 15;4:88. doi: 10.3389/fimmu.2013.00088. eCollection 2013. Front Immunol. 2013. PMID: 23596443 Free PMC article.
References
- Barr FA, Puype M, Vandekerckhove J, Warren G (1997) GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91: 253–262 - PubMed
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144: 253–267 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI069345/AI/NIAID NIH HHS/United States
- R37 AI042999/AI/NIAID NIH HHS/United States
- RC1AI086845/AI/NIAID NIH HHS/United States
- R01 AI042999/AI/NIAID NIH HHS/United States
- RC1 AI086845/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases