Recent loss of vitamin C biosynthesis ability in bats - PubMed (original) (raw)
Recent loss of vitamin C biosynthesis ability in bats
Jie Cui et al. PLoS One. 2011.
Abstract
The traditional assumption that bats cannot synthesize vitamin C (Vc) has been challenged recently. We have previously shown that two Old World bat species (Rousettus leschenaultii and Hipposideros armiger) have functional L-gulonolactone oxidase (GULO), an enzyme that catalyzes the last step of Vc biosynthesis de novo. Given the uncertainties surrounding when and how bats lost GULO function, exploration of gene evolutionary patterns is needed. We therefore sequenced GULO genes from 16 bat species in 5 families, aiming to establish their evolutionary histories. In five cases we identified pseudogenes for the first time, including two cases in the genus Pteropus (P. pumilus and P. conspicillatus) and three in family Hipposideridae (Coelops frithi, Hipposideros speoris, and H. bicolor). Evolutionary analysis shows that the Pteropus clade has the highest ω ratio and has been subjected to relaxed selection for less than 3 million years. Purifying selection acting on the pseudogenized GULO genes of roundleaf bats (family Hipposideridae) suggests they have lost the ability to synthesize Vc recently. Limited mutations in the reconstructed GULO sequence of the ancestor of all bats contrasts with the many mutations in the ancestral sequence of recently emerged Pteropus bats. We identified at least five mutational steps that were then related to clade origination times. Together, our results suggest that bats lost the ability to biosynthesize vitamin C recently by exhibiting stepwise mutation patterns during GULO evolution that can ultimately lead to pseudogenization.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Alignments of bat GULO nucleotide gene sequences.
(A) The intact bat GULO nucleotide gene sequences; (B) the bat GULO pseudogenized nucleotide gene sequences. The nucleotide position numbers are denoted according to the nucleotide sequence of Rousettus leschenaultii GULO (HQ415789). The boxes donate insertions, deletions, or premature stop codons that break the gene reading frames.
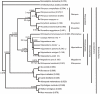
Figure 2. Phylogenetic tree based on GULO genes.
The evolutionary history was reconstructed using the NJ method in MEGA4 . The bootstrap consensus tree inferred from 2,000 replicates is shown to represent GULO evolution for each taxon. Bootstrap values lower than 50% are no shown. The scale bar represents genetic distance. The evolutionary distances were computed using the Maximum Composite Likelihood model. Lineages for pseudogenes are marked with ψ.
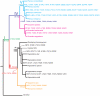
Figure 3. Selection pressures acting on GULO genes.
The published species tree , is shown and selection pressures marked were calculated using the free-ratio model in PAML4 . Values given on the branches, or in parentheses, are ω ratios (dN/dS) estimated by maximum-likelihood. Values of infinity (∞, dS = 0) are not shown. The time of origin for each ancestral node was collected from published data , . Lineages for pseudogenes are marked with ψ.
Figure 4. Stepwise mutation patterns during bat GULO evolution.
Amino acid (abbreviation) changes of bat GULO are shown after the divergence of bats with non-bat Laurasiatheria species. The topology was retrieved from published species trees , . Five major steps were identified according to the mutation pattern, and they are highlighted in Roman characters (I, II, III, IV, and V). The time of origin for each node was collected from published data , . The positions of mutations are recorded according to the protein sequence of the Rousettus leschenaultii GULO gene (ADP88813). For example, G119A means that the amino acid G evolved from A at amino acid position 119. The outgroup of non-bat Laurasiatheria species include pig, cow, horse, cat, and panda.
Similar articles
- Progressive pseudogenization: vitamin C synthesis and its loss in bats.
Cui J, Pan YH, Zhang Y, Jones G, Zhang S. Cui J, et al. Mol Biol Evol. 2011 Feb;28(2):1025-31. doi: 10.1093/molbev/msq286. Epub 2010 Oct 29. Mol Biol Evol. 2011. PMID: 21037206 - Conserved or lost: molecular evolution of the key gene GULO in vertebrate vitamin C biosynthesis.
Yang H. Yang H. Biochem Genet. 2013 Jun;51(5-6):413-25. doi: 10.1007/s10528-013-9574-0. Epub 2013 Feb 12. Biochem Genet. 2013. PMID: 23404229 - Functional rescue of vitamin C synthesis deficiency in human cells using adenoviral-based expression of murine l-gulono-gamma-lactone oxidase.
Ha MN, Graham FL, D'Souza CK, Muller WJ, Igdoura SA, Schellhorn HE. Ha MN, et al. Genomics. 2004 Mar;83(3):482-92. doi: 10.1016/j.ygeno.2003.08.018. Genomics. 2004. PMID: 14962674 - Molecular basis for the deficiency in humans of gulonolactone oxidase, a key enzyme for ascorbic acid biosynthesis.
Nishikimi M, Yagi K. Nishikimi M, et al. Am J Clin Nutr. 1991 Dec;54(6 Suppl):1203S-1208S. doi: 10.1093/ajcn/54.6.1203s. Am J Clin Nutr. 1991. PMID: 1962571 Review. - L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis.
Gad AAM, Sirko A. Gad AAM, et al. Curr Issues Mol Biol. 2024 Oct 1;46(10):11057-11074. doi: 10.3390/cimb46100657. Curr Issues Mol Biol. 2024. PMID: 39451537 Free PMC article. Review.
Cited by
- Multiple independent L-gulonolactone oxidase (GULO) gene losses and vitamin C synthesis reacquisition events in non-Deuterostomian animal species.
Henriques SF, Duque P, López-Fernández H, Vázquez N, Fdez-Riverola F, Reboiro-Jato M, Vieira CP, Vieira J. Henriques SF, et al. BMC Evol Biol. 2019 Jun 18;19(1):126. doi: 10.1186/s12862-019-1454-8. BMC Evol Biol. 2019. PMID: 31215418 Free PMC article. - A "forward genomics" approach links genotype to phenotype using independent phenotypic losses among related species.
Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G. Hiller M, et al. Cell Rep. 2012 Oct 25;2(4):817-23. doi: 10.1016/j.celrep.2012.08.032. Epub 2012 Sep 27. Cell Rep. 2012. PMID: 23022484 Free PMC article. - Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes.
Wheeler G, Ishikawa T, Pornsaksit V, Smirnoff N. Wheeler G, et al. Elife. 2015 Mar 13;4:e06369. doi: 10.7554/eLife.06369. Elife. 2015. PMID: 25768426 Free PMC article. - Molecular evolution of anthocyanin pigmentation genes following losses of flower color.
Ho WW, Smith SD. Ho WW, et al. BMC Evol Biol. 2016 May 10;16:98. doi: 10.1186/s12862-016-0675-3. BMC Evol Biol. 2016. PMID: 27161359 Free PMC article. - Vitamin C deficiency reveals developmental differences between neonatal and adult hematopoiesis.
Phadke I, Pouzolles M, Machado A, Moraly J, Gonzalez-Menendez P, Zimmermann VS, Kinet S, Levine M, Violet PC, Taylor N. Phadke I, et al. Front Immunol. 2022 Sep 30;13:898827. doi: 10.3389/fimmu.2022.898827. eCollection 2022. Front Immunol. 2022. PMID: 36248829 Free PMC article.
References
- Linster CL, Van Schaftingen E. Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. - PubMed
- Chatterjee IB, Kar NC, Ghosh NC, Guha BC. Aspects of ascorbic acid biosynthesis in animals. Ann N Y Acad Sci. 1961;92:36–56. - PubMed
- Troadec MB, Kaplan J. Some vertebrates go with the GLO. Cell. 2008;132:921–922. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical