Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation - PubMed (original) (raw)
Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation
Maria L Mouchess et al. Immunity. 2011.
Abstract
Recognition of nucleic acids as a signature of infection by Toll-like receptors (TLRs) 7 and 9 exposes the host to potential self-recognition and autoimmunity. It has been proposed that intracellular compartmentalization is largely responsible for reliable self versus nonself discrimination by these receptors. We have previously shown that TLR9 and TLR7 require processing prior to activation, which may further reinforce receptor compartmentalization and tolerance to self, yet this possibility remains untested. Here we report that residues within the TLR9 transmembrane (TM) region conferred the requirement for ectodomain proteolysis. TLR9 TM mutants responded to extracellular DNA, and mice expressing such receptors died from systemic inflammation and anemia. This inflammatory disease did not require lymphocytes and appeared to require recognition of self-DNA by dendritic cells. To our knowledge, these results provide the first demonstration that TLR-intrinsic mutations can lead to a break in tolerance.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
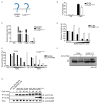
Figure 1. TLR9TM-MUT does not require processing for activation
A, Schematic of TLR9 and TLR9TM-MUT. TLR9TM-MUT consists of the ectodomain and cytosolic domain of TLR9 (blue) and the transmembrane domain of TLR3 (yellow). B, TLR9TM-MUT can respond to CpG DNA ligands. HEK293 cells stably expressing a NF-κB luciferase reporter and TLR9 or TLR9TM-MUT were stimulated with CpG ODN (phosphorothioate backbone) as indicated and luciferase activity was measured 16hr later. C, Macrophages expressing TLR9TM-MUT respond to CpG ODN and extracellular genomic DNA. TLR9-deficient macrophages transduced with retroviruses encoding TLR9, TLR9TM-MUT or empty vector (control) were stimulated as indicated and the percentage of GFP-positive cells producing TNF was measured by intracellular staining and flow cytometry. Data shown are representative of two experiments. D and E, Signaling by TLR9TM-MUT does not require proteolysis. TLR9-deficient macrophages expressing TLR9 or TLR9TM-MUT were pretreated with bafilomycinA1 for 2 hours (D) or Z-FA-FMK overnight (E), stimulated with CpG ODN, and stained to measure TNF production, as described in (C). F, Stabilization of shifted form of TLR9TM-MUT in the presence of Z-FA-FMK. Anti-HA immunoblot of TLR9 and TLR9TM-MUT from lysates of macrophage treated with DMSO (vehicle) or Z-FA-FMK. Open triangle indicates full-length TLR9. Asterisk denotes the shifted form of the full-length receptor. Immunoblot shown is representative of two experiments. G, Full-length TLR9TM-MUT can recruit MyD88. TLR9-deficient macrophages expressing N-terminal FLAG-TLR9 or FLAG-TLR9TM-MUT were stimulated with CpG ODN and recruitment of MyD88 was measured by immunoprecipitation (IP) and immunoblot (IB) as indicated. The immunoblot of whole cell lysates (WCL) shows equivalent amounts of MyD88. Data shown are representative of two experiments. Unless stated otherwise, experiments in this figure were performed at least three times with similar results.
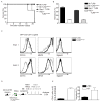
Figure 2. TLR9TM-MUT can access the cell surface and responds to extracellular ligands
A, TLR9TM-MUT is more responsive to extracellular DNA ligands. Luciferase assays were performed as described in (B) except stimulations were with either phosphodiester (PD) backbone CpG ODN or salmon sperm DNA (V. DNA). Data shown are representative of two experiments. B, TLR9TM-MUT can respond to immobilized CpG ODN at the cell surface. Cells described in (Fig.1B) were plated on streptavidin coated plates to which biotinylated-CpG ODN had been previously conjugated. Luciferase activity was measured in lysates after 6h. C, Full-length TLR9TM-MUT is expressed at the cell surface. Flow cytometry histograms of anti-FLAG staining of HEK293T cells stably expressing N-terminal FLAG-TLR9 (grey), FLAG-TLR9TM-MUT (black), FLAG-TLR2 (blue), or empty vector control (shaded). Anti-FLAG immunoblot of whole cell lysates (WCL) of the indicated cells (inset). See also Figure S2A. D, The ectodomain of TLR9TM-MUT is processed inefficiently. Anti-HA immunoblot of lysates from TLR9-deficient macrophages expressing C-terminally HA-tagged TLR9 or TLR9TM-MUT. Open triangle indicates full-length TLR9. Closed triangle indicates processed TLR9. Asterisk denotes the shifted form of the full-length receptor. E, TLR9TM-MUT traffics through the Golgi apparatus. Immunoprecipitated TLR9 or TLR9TM-MUT from macrophage lysates were treated with Endoglycosidase H (E), PNGase F (P) or left untreated (U). F, TLR9 and TLRTM-MUT exit the ER with similar efficiency and kinetics. Pulse-chase analysis of macrophages expressing TLR9 or TLR9TM-MUT. TLR9 or TLR9TM-MUT were harvested at the indicated chase times, immunoprecipitated and visualized by SDS-PAGE. Data presented are representative of at least three independent experiments.
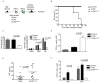
Figure 3. Expression of TLR9TM-MUT in vivo leads to fatal inflammation
A, Schematic of the approach used to express TLR9 or TLR9TM-MUT in mice. Hematopoietic stem cells (HSC) from TLR9-deficient mice were transduced with retroviruses encoding TLR9, TLR9TM-MUT, or vector control followed by transfer into lethally irradiated C57BL/6 recipient mice. B, Mice expressing TLR9TM-MUT die from inflammatory disease. Survival plot of radiation chimeric mice receiving HSCs expressing either TLR9, TLR9TM-MUT, or empty vector (control). C, TLR9TM-MUT expressing mice develop anemia. Mean values of hematocrit, red blood cells (RBC), and hemoglobin (Hg) in the blood of mice at day 19 post-HSC transfer. D, Expression of TLR9TM-MUT in mice causes an expansion of DCs. Graph showing the total number of GFP-positive, CD11c+ cells in the bone marrow at day 19 after injection of HSCs. E, TLR9TM-MUT expression leads to inflammatory cytokine induction. TNF was measured in sera from the indicated mice at day 19-29 post-HSC transfer. Each point represents an individual mouse. Horizontal line represents the mean. F, Expression of TLR9TM-MUT in mice leads to block in B cell development. The total number of CD19+ cells in the bone marrow and spleens of the indicated mice, as measured by flow cytometry, is shown. Unless stated otherwise, experiments in this figure were performed at least three times with similar results. See also Figure S3.
Figure 4. TLR9TM-MUT mediated inflammatory disease is driven by CD11c+ dendritic cells
A and B, Disease caused by TLR9TM-MUT does not require B or T cells. Experiments were performed as in (3B-D), except Rag1-deficient HSCs were used to generate chimeric mice. A, Survival plot of chimeric mice receiving transduced HSCs of the indicated genotypes. B, Blood analyses at day 19 post-HSC transfer are shown. The data shown are representative of two independent experiments. C, Surface phenotype of CD11c+ cells in TLR9TM-MUT radiation chimeras. Bone marrow cells from radiation chimeras expressing the indicated receptors were analyzed for cell surface marker expression by flow cytometry (d20 post-transfer). The histograms show expression of the CD11b, MHC class II, and SiglecH on GFP-gated CD11c+ cells. Representative histograms are shown for individual mice in two separate experiments containing four mice. D and E, CD11c+ cells are required for inflammatory disease in TLR9TM-MUT mice. D, Schematic of experimental setup for depletion of CD11c+ cells in radiation chimeras expressing TLR9TM-MUT. Mice were given either PBS and Diphtheria Toxin (DT) every five days after introduction of TLR9TM-MUT transduced HSCs and analyzed twenty days after HSC injection. E, Block in B cell development and anemia seen in TLR9TM-MUT radiation chimeras is rescued by depletion of CD11c+ cells. The total number of CD19+ cells in the spleens of the indicated mice as well as mean hematocrit are shown for PBS or Diphtheria toxin (DT)-treated mice. Data are representative of two experiments with four mice per group. See also Figure S4A and B.
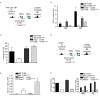
Figure 5. Lack of TNF and lymphotoxin αβ but not IFNAR partially rescues inflammatory disease in TLR9TM-MUT expressing mice
A-C, Autoinflammatory disease mediated by TLR9TM-MUT is partially ameliorated in the absence of tnf and lymphotoxinαß. A, Schematic of bone marrow chimera experiment to address role of TNF in TLR9TM-MUT mediated disease. B, Partial recovery of B cell development in TLR9TM-MUT expressing radiation chimeras generated with Tnfa-/-Lta-/-Ltb-/- HSCs. The number of CD19+ B cells in the bone marrow (BM) and spleen (SPL) of TLR9 and TLR9TM-MUT expressing radiation chimeras in C57BL/6 or Tnfa-/-Lta-/-Ltb-/- cells (d20 post-transfer) is shown. C, Tnfa-/-Lta-/-Ltb-/- TLR9TM-MUT expressing mice do not develop anemia at early time points. Mean values of hematocrit in the blood of mice at day 20 post-HSC transfer. Data shown are representative of two experiments. D-F, Loss of Ifnar1 does not rescue TLR9TM-MUT mediated disease in vivo. D, Schematic of bone marrow chimera experiment to address role of type I interferon in TLR9TM-MUT mediated disease. Initial experiment utilized IFNAR-deficient or C57BL/6 donors. E, Expression of TLR9TM-MUT using IFNAR-deficient HSCs in mice causes an expansion of dendritic cells similar to wild type. Graph showing the average number of GFP-gated CD11c+ cells in the bone marrow and spleen at day 25 after injection of HSCs. F, TLR9TM-MUT expression in IFNAR-deficient HSCs expressing mice develop severe anemia. Mean values of hematocrit, red blood cells (RBC), and hemoglobin (Hg) in the blood at day 25 post-HSC transfer. See also Figure S5.
Figure 6. TLR9 TM mutants identify residues critical in preventing self-reactivity
A, Schematic showing an alignment of TM regions of TLR9 mutants. Underline indicates residues from TLR9. Arrowhead indicates the last conserved cysteine of the ectodomain. B, TLR9TM-MUT4 has altered trafficking to the endolysosome. Anti-HA immunoblots of lysates from TLR9-deficient macrophages expressing the indicated TLR9 TM mutants, as described in (A). C, TLR9TM-MUT4 can respond to ligands at the cell surface. HEK293 cells stably expressing TLR9 or the indicated TLR9 mutants were plated on streptavidin-coated plates to which biotinylated CpG ODN had been previously conjugated. Luciferase activity was measured in lysates after 6hr. D, TLR9TM-MUT4 activation does not require proteolysis. TLR9-deficient macrophages expressing the indicated TLR9 TM mutants were stimulated with CpG ODN after treatment with DMSO (vehicle) or Z-FA-FMK. TNF production was measured by flow cytometry. The data are normalized for each TLR9 TM mutant by dividing by the percentage of responding cells in vehicle treated control samples. Results shown for B-D are representative of at least three experiments. E, TLR9TM-MUT4 induces inflammatory disease in vivo. Mean values of hematocrit, red blood cells (RBC), and hemoglobin (Hg) in the blood of mice at day 19 post-HSC transfer. Data are representative of two independent experiments.
Comment in
- Innate immunity: TLR9 mutations reveal a new level of self tolerance.
Papatriantafyllou M. Papatriantafyllou M. Nat Rev Immunol. 2011 Dec 9;12(1):7. doi: 10.1038/nri3134. Nat Rev Immunol. 2011. PMID: 22158413 No abstract available.
Similar articles
- The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor.
Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. Ewald SE, et al. Nature. 2008 Dec 4;456(7222):658-62. doi: 10.1038/nature07405. Epub 2008 Sep 28. Nature. 2008. PMID: 18820679 Free PMC article. - B cell autophagy mediates TLR7-dependent autoimmunity and inflammation.
Weindel CG, Richey LJ, Bolland S, Mehta AJ, Kearney JF, Huber BT. Weindel CG, et al. Autophagy. 2015;11(7):1010-24. doi: 10.1080/15548627.2015.1052206. Autophagy. 2015. PMID: 26120731 Free PMC article. - Tyrosine 870 of TLR9 is critical for receptor maturation rather than phosphorylation-dependent ligand-induced signaling.
Biswas C, Rao S, Slade K, Hyman D, Dersh D, Mantegazza AR, Zoltick PW, Marks MS, Argon Y, Behrens EM. Biswas C, et al. PLoS One. 2018 Jul 19;13(7):e0200913. doi: 10.1371/journal.pone.0200913. eCollection 2018. PLoS One. 2018. PMID: 30024926 Free PMC article. - Regulation of TLR7/9 signaling in plasmacytoid dendritic cells.
Bao M, Liu YJ. Bao M, et al. Protein Cell. 2013 Jan;4(1):40-52. doi: 10.1007/s13238-012-2104-8. Epub 2012 Nov 7. Protein Cell. 2013. PMID: 23132256 Free PMC article. Review. - Regulation of dendritic cell function through Toll-like receptors.
Kaisho T, Akira S. Kaisho T, et al. Curr Mol Med. 2003 Jun;3(4):373-85. doi: 10.2174/1566524033479726. Curr Mol Med. 2003. PMID: 12776992 Review.
Cited by
- Nucleic Acid Sensing by Toll-Like Receptors in the Endosomal Compartment.
Miyake K, Shibata T, Fukui R, Sato R, Saitoh SI, Murakami Y. Miyake K, et al. Front Immunol. 2022 Jun 23;13:941931. doi: 10.3389/fimmu.2022.941931. eCollection 2022. Front Immunol. 2022. PMID: 35812450 Free PMC article. Review. - Large-scale mutational analysis identifies UNC93B1 variants that drive TLR-mediated autoimmunity in mice and humans.
Rael VE, Yano JA, Huizar JP, Slayden LC, Weiss MA, Turcotte EA, Terry JM, Zuo W, Thiffault I, Pastinen T, Farrow EG, Jenkins JL, Becker ML, Wong SC, Stevens AM, Otten C, Allenspach EJ, Bonner DE, Bernstein JA, Wheeler MT, Saxton RA; Undiagnosed Diseases Network; Liu B, Majer O, Barton GM. Rael VE, et al. J Exp Med. 2024 Aug 5;221(8):e20232005. doi: 10.1084/jem.20232005. Epub 2024 May 23. J Exp Med. 2024. PMID: 38780621 Free PMC article. - Red Blood Cells Homeostatically Bind Mitochondrial DNA through TLR9 to Maintain Quiescence and to Prevent Lung Injury.
Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, Rivella S, Worthen GS, Mangalmurti NS. Hotz MJ, et al. Am J Respir Crit Care Med. 2018 Feb 15;197(4):470-480. doi: 10.1164/rccm.201706-1161OC. Am J Respir Crit Care Med. 2018. PMID: 29053005 Free PMC article. - TLR9 re-expression in cancer cells extends the S-phase and stabilizes p16(INK4a) protein expression.
Parroche P, Roblot G, Le Calvez-Kelm F, Tout I, Marotel M, Malfroy M, Durand G, McKay J, Ainouze M, Carreira C, Allatif O, Traverse-Glehen A, Mendiola M, Pozo-Kreilinger JJ, Caux C, Tommasino M, Goutagny N, Hasan UA. Parroche P, et al. Oncogenesis. 2016 Jul 25;5(7):e244. doi: 10.1038/oncsis.2016.49. Oncogenesis. 2016. PMID: 27454079 Free PMC article. - Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes.
Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, Coyle AJ, Kolbeck R, Green DR, Sanjuan MA. Henault J, et al. Immunity. 2012 Dec 14;37(6):986-997. doi: 10.1016/j.immuni.2012.09.014. Epub 2012 Dec 6. Immunity. 2012. PMID: 23219390 Free PMC article.
References
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. - PubMed
- Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature. 1985;318:667–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI072429-02S1/AI/NIAID NIH HHS/United States
- AI072429/AI/NIAID NIH HHS/United States
- F31AI083012/AI/NIAID NIH HHS/United States
- F31 AI083012/AI/NIAID NIH HHS/United States
- R01 AI072429-01A2/AI/NIAID NIH HHS/United States
- R01 AI072429/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases