eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges - PubMed (original) (raw)
. 2012 Jan;40(Database issue):D284-9.
doi: 10.1093/nar/gkr1060. Epub 2011 Nov 16.
Damian Szklarczyk, Kalliopi Trachana, Alexander Roth, Michael Kuhn, Jean Muller, Roland Arnold, Thomas Rattei, Ivica Letunic, Tobias Doerks, Lars J Jensen, Christian von Mering, Peer Bork
Affiliations
- PMID: 22096231
- PMCID: PMC3245133
- DOI: 10.1093/nar/gkr1060
eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges
Sean Powell et al. Nucleic Acids Res. 2012 Jan.
Abstract
Orthologous relationships form the basis of most comparative genomic and metagenomic studies and are essential for proper phylogenetic and functional analyses. The third version of the eggNOG database (http://eggnog.embl.de) contains non-supervised orthologous groups constructed from 1133 organisms, doubling the number of genes with orthology assignment compared to eggNOG v2. The new release is the result of a number of improvements and expansions: (i) the underlying homology searches are now based on the SIMAP database; (ii) the orthologous groups have been extended to 41 levels of selected taxonomic ranges enabling much more fine-grained orthology assignments; and (iii) the newly designed web page is considerably faster with more functionality. In total, eggNOG v3 contains 721,801 orthologous groups, encompassing a total of 4,396,591 genes. Additionally, we updated 4873 and 4850 original COGs and KOGs, respectively, to include all 1133 organisms. At the universal level, covering all three domains of life, 101,208 orthologous groups are available, while the others are applicable at 40 more limited taxonomic ranges. Each group is amended by multiple sequence alignments and maximum-likelihood trees and broad functional descriptions are provided for 450,904 orthologous groups (62.5%).
Figures
Figure 1.
In addition to the over 100 000 orthologous groups in the last universal common ancestor (LUCA), eggNOG v3 also provides orthologous groups and functional annotation for an additional 40 taxonomic levels. Here we display each level with its abbreviated name, species count, orthologous group count and annotation coverage. The annotation coverage for both the functional description of the groups as well as the functional category (in parentheses) is given.
Figure 2.
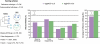
Quality assessment of eggNOG v3. We used 70 manually curated families (RefOGs) to test the accuracy of orthology prediction of the new release compared to eggNOG v2. For each release, we identified the orthologous group (OG) with the largest overlap of each RefOG and calculated how many genes were not predicted in the OG (missing orthologs) and how many genes were over-predicted in the OG (false assignments). Additionally, we checked if members of the same RefOG have been separated into multiple OGs (RefOG fission) and how many of those OGs include more than three false assignments (RefOG fusion). Missing orthologs influence 41% of the RefOGs; however, this is significantly less than the 57% in eggNOG v2. Similarly, less RefOGs include false assignments in eggNOG v3 (60%) compared to version 2 (66%). However, there are slightly less artificial OG fusions and fissions in eggNOG v2. Given that an addition of species can introduce false assignments, our results suggest that the eggNOG methodology can tolerate a large number of species, and at the same time improve its coverage against the tested benchmark dataset.
Figure 3.
Screenshot of a results page. The eggNOG database was queried for the term ‘smoothened’. The top left picture demonstrates the simplified navigation of multiple search terms and species selection. The navigation tree at the top right of the page allows the user to change the view to more coarse-grained orthologous groups, for example, the mammalian orthologous groups. The group features, such as member proteins, alignments (green arrow) and phylogenetic trees with SMART domains (orange arrow), can be accessed inline and do not require a page refresh.
Similar articles
- eggNOG: automated construction and annotation of orthologous groups of genes.
Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. Jensen LJ, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D250-4. doi: 10.1093/nar/gkm796. Epub 2007 Oct 16. Nucleic Acids Res. 2008. PMID: 17942413 Free PMC article. - eggNOG 6.0: enabling comparative genomics across 12 535 organisms.
Hernández-Plaza A, Szklarczyk D, Botas J, Cantalapiedra CP, Giner-Lamia J, Mende DR, Kirsch R, Rattei T, Letunic I, Jensen LJ, Bork P, von Mering C, Huerta-Cepas J. Hernández-Plaza A, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D389-D394. doi: 10.1093/nar/gkac1022. Nucleic Acids Res. 2023. PMID: 36399505 Free PMC article. - eggNOG v4.0: nested orthology inference across 3686 organisms.
Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldón T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. Powell S, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D231-9. doi: 10.1093/nar/gkt1253. Epub 2013 Dec 1. Nucleic Acids Res. 2014. PMID: 24297252 Free PMC article. - eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations.
Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, Bork P. Muller J, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D190-5. doi: 10.1093/nar/gkp951. Epub 2009 Nov 9. Nucleic Acids Res. 2010. PMID: 19900971 Free PMC article. - The quest for orthologs: finding the corresponding gene across genomes.
Kuzniar A, van Ham RC, Pongor S, Leunissen JA. Kuzniar A, et al. Trends Genet. 2008 Nov;24(11):539-51. doi: 10.1016/j.tig.2008.08.009. Epub 2008 Sep 24. Trends Genet. 2008. PMID: 18819722 Review.
Cited by
- CoPAP: Coevolution of presence-absence patterns.
Cohen O, Ashkenazy H, Levy Karin E, Burstein D, Pupko T. Cohen O, et al. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W232-7. doi: 10.1093/nar/gkt471. Epub 2013 Jun 8. Nucleic Acids Res. 2013. PMID: 23748951 Free PMC article. - Orthologous gene clusters and taxon signature genes for viruses of prokaryotes.
Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. Kristensen DM, et al. J Bacteriol. 2013 Mar;195(5):941-50. doi: 10.1128/JB.01801-12. Epub 2012 Dec 7. J Bacteriol. 2013. PMID: 23222723 Free PMC article. - Transcriptome sequences spanning key developmental states as a resource for the study of the cestode Schistocephalus solidus, a threespine stickleback parasite.
Hébert FO, Grambauer S, Barber I, Landry CR, Aubin-Horth N. Hébert FO, et al. Gigascience. 2016 Jun 2;5:24. doi: 10.1186/s13742-016-0128-3. Gigascience. 2016. PMID: 27259971 Free PMC article. - Transcriptome profiling of trichome-less reveals genes associated with multicellular trichome development in Cucumis sativus.
Zhao JL, Wang YL, Yao DQ, Zhu WY, Chen L, He HL, Pan JS, Cai R. Zhao JL, et al. Mol Genet Genomics. 2015 Oct;290(5):2007-18. doi: 10.1007/s00438-015-1057-z. Epub 2015 May 8. Mol Genet Genomics. 2015. PMID: 25952908 - Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome.
Omasits U, Quebatte M, Stekhoven DJ, Fortes C, Roschitzki B, Robinson MD, Dehio C, Ahrens CH. Omasits U, et al. Genome Res. 2013 Nov;23(11):1916-27. doi: 10.1101/gr.151035.112. Epub 2013 Jul 22. Genome Res. 2013. PMID: 23878158 Free PMC article.
References
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. - PubMed
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. - PubMed
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. - PubMed
- Huynen MA, Snel B, von Mering C, Bork P. Function prediction and protein networks. CuCrr. Opin. Cell. Biol. 2003;15:191–198. - PubMed