Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa - PubMed (original) (raw)
. 2011 Nov;17(11):2113-21.
doi: 10.3201/eid1711.110794.
Gary Van Domselaar, Steven Stroika, Matthew Walker, Heather Kent, Cheryl Tarr, Deborah Talkington, Lori Rowe, Melissa Olsen-Rasmussen, Michael Frace, Scott Sammons, Georges Anicet Dahourou, Jacques Boncy, Anthony M Smith, Philip Mabon, Aaron Petkau, Morag Graham, Matthew W Gilmour, Peter Gerner-Smidt; V. cholerae Outbreak Genomics Task Force
Collaborators, Affiliations
- PMID: 22099115
- PMCID: PMC3310578
- DOI: 10.3201/eid1711.110794
Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa
Aleisha R Reimer et al. Emerg Infect Dis. 2011 Nov.
Abstract
Cholera was absent from the island of Hispaniola at least a century before an outbreak that began in Haiti in the fall of 2010. Pulsed-field gel electrophoresis (PFGE) analysis of clinical isolates from the Haiti outbreak and recent global travelers returning to the United States showed indistinguishable PFGE fingerprints. To better explore the genetic ancestry of the Haiti outbreak strain, we acquired 23 whole-genome Vibrio cholerae sequences: 9 isolates obtained in Haiti or the Dominican Republic, 12 PFGE pattern-matched isolates linked to Asia or Africa, and 2 nonmatched outliers from the Western Hemisphere. Phylogenies for whole-genome sequences and core genome single-nucleotide polymorphisms showed that the Haiti outbreak strain is genetically related to strains originating in India and Cameroon. However, because no identical genetic match was found among sequenced contemporary isolates, a definitive genetic origin for the outbreak in Haiti remains speculative.
Figures
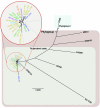
Figure 1
Whole-genome neighbor-joining tree of Vibrio cholerae isolate from cholera outbreak in Haiti, fall 2010; concurrent clinical isolates with pulsed-field gel electrophoresis pattern-matched combinations; reference isolates sequenced in this study; and available reference sequences. Sequence alignments of quality draft or complete genomes were performed by using Progressive Mauve (16) and visualized by using PhyML version 3.0 (17). Whole-genome relationship of Haiti isolates with closest genetic relatives is shown in the inset. Scale bar indicates nucleotides substitutions per site.
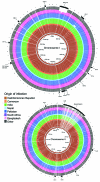
Figure 2
BLAST (
http://blast.ncbi.nlm.gov/Blast.cgi
) atlas of predicted protein homologies mapped against the closed genome of Haiti Vibrio cholerae outbreak type isolate 2010EL-1786, fall 2010. Full color saturation represents 100% sequence homology, and gaps indicate regions of divergence. Gaps in the innermost (red) circle for reference isolate 2010EL-1786 represent gaps between coding sequences, not genetic diversity. A) Chromosome I; B) chromosome II. From center: Haiti/Dominican Republic isolates 2010EL-1786, 2010EL-1961, 2011EL-1089, 2010EL-2010N, 2010EL-2010H, 2011V-1021, 2010EL-1798, 2010EL-1792, and 2011EL-1133; Cameroon isolate 2010EL-1749; India isolates 2009V-1085, 2009V-1096, 2009V-1131, 3546–06, and 3500–05; Nepal isolate 3554–08; Pakistan isolates 3582–05, 2009V-1046, 2010-V1014, and 2009V-1116; South Africa isolate 2011EL-1137; Bangladesh isolates CIRS101, MJ-1236 and N16961; and other isolates C6706, M66–2, and 3569–08.
Figure 3
Genetic structure of cholera toxin (CTX) prophage and associated elements in Haiti cholera outbreak Vibrio cholerae isolate 2010EL-1786, fall 2010. The toxin-linked cryptic (TLC) element is not drawn to scale. Black arrows indicate the direction of transcription for each coding region. Red, forward transcription; blue, reverse transcription; gray, predicted open reading frame with no experimental evidence.
Similar articles
- Characterization of toxigenic Vibrio cholerae from Haiti, 2010-2011.
Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Gerner-Smidt P. Talkington D, et al. Emerg Infect Dis. 2011 Nov;17(11):2122-9. doi: 10.3201/eid1711.110805. Emerg Infect Dis. 2011. PMID: 22099116 Free PMC article. - Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak.
Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. Hendriksen RS, et al. mBio. 2011 Sep 1;2(4):e00157-11. doi: 10.1128/mBio.00157-11. Print 2011. mBio. 2011. PMID: 21862630 Free PMC article. - The origin of the Haitian cholera outbreak strain.
Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. Chin CS, et al. N Engl J Med. 2011 Jan 6;364(1):33-42. doi: 10.1056/NEJMoa1012928. Epub 2010 Dec 9. N Engl J Med. 2011. PMID: 21142692 Free PMC article. - Widespread epidemic cholera caused by a restricted subset of Vibrio cholerae clones.
Moore S, Thomson N, Mutreja A, Piarroux R. Moore S, et al. Clin Microbiol Infect. 2014 May;20(5):373-9. doi: 10.1111/1469-0691.12610. Epub 2014 Mar 29. Clin Microbiol Infect. 2014. PMID: 24575898 Review. - Genomic science in understanding cholera outbreaks and evolution of Vibrio cholerae as a human pathogen.
Robins WP, Mekalanos JJ. Robins WP, et al. Curr Top Microbiol Immunol. 2014;379:211-29. doi: 10.1007/82_2014_366. Curr Top Microbiol Immunol. 2014. PMID: 24590676 Free PMC article. Review.
Cited by
- Review of molecular subtyping methodologies used to investigate outbreaks due to multidrug-resistant enteric bacterial pathogens in sub-Saharan Africa.
Smith AM. Smith AM. Afr J Lab Med. 2019 Mar 14;8(1):760. doi: 10.4102/ajlm.v8i1.760. eCollection 2019. Afr J Lab Med. 2019. PMID: 31205868 Free PMC article. Review. - Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India.
Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. Ghosh P, et al. PLoS One. 2014 Nov 21;9(11):e112973. doi: 10.1371/journal.pone.0112973. eCollection 2014. PLoS One. 2014. PMID: 25415339 Free PMC article. - Food Safety in the Age of Next Generation Sequencing, Bioinformatics, and Open Data Access.
Taboada EN, Graham MR, Carriço JA, Van Domselaar G. Taboada EN, et al. Front Microbiol. 2017 May 23;8:909. doi: 10.3389/fmicb.2017.00909. eCollection 2017. Front Microbiol. 2017. PMID: 28588568 Free PMC article. Review. - Genomic diversity of 2010 Haitian cholera outbreak strains.
Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. Hasan NA, et al. Proc Natl Acad Sci U S A. 2012 Jul 17;109(29):E2010-7. doi: 10.1073/pnas.1207359109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711841 Free PMC article. - Sequence polymorphisms of rfbT among the Vibrio cholerae O1 strains in the Ogawa and Inaba serotype shifts.
Liang W, Wang L, Liang P, Zheng X, Zhou H, Zhang J, Zhang L, Kan B. Liang W, et al. BMC Microbiol. 2013 Jul 26;13:173. doi: 10.1186/1471-2180-13-173. BMC Microbiol. 2013. PMID: 23889924 Free PMC article.
References
- Pan American Health Organization. Cholera and post-earthquake response in Haiti, 2011. Jul 25 [cited 2011 Aug 6]. http://www.who.int/hac/crises/hti/sitreps/haiti_health_cluster_bulletin_....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases