Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment - PubMed (original) (raw)
Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment
Yuzhen Ye. Proceedings (IEEE Int Conf Bioinformatics Biomed). 2011.
Abstract
16S rRNA gene profiling has recently been boosted by the development of pyrosequencing methods. A common analysis is to group pyrosequences into Operational Taxonomic Units (OTUs), such that reads in an OTU are likely sampled from the same species. However, species diversity estimated from error-prone 16S rRNA pyrosequences may be inflated because the reads sampled from the same 16S rRNA gene may appear different, and current OTU inference approaches typically involve time-consuming pairwise/multiple distance calculation and clustering. I propose a novel approach AbundantOTU based on a Consensus Alignment (CA) algorithm, which infers consensus sequences, each representing an OTU, taking advantage of the sequence redundancy for abundant species. Pyrosequencing reads can then be recruited to the consensus sequences to give quantitative information for the corresponding species. As tested on 16S rRNA pyrosequence datasets from mock communities with known species, AbundantOTU rapidly reported identified sequences of the source 16S rRNAs and the abundances of the corresponding species. AbundantOTU was also applied to 16S rRNA pyrosequence datasets derived from real microbial communities and the results are in general agreement with previous studies.
Figures
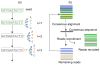
Figure 1
A schematic demonstration of AbundantOTU algorithm by using consensus alignment. (a) Consensus alignment by using a dynamic programming algorithm, adding one nucleotide at a time. (b) Abundant OTU inference by deriving consensus sequence and recruiting reads to the consensus sequence iteratively.
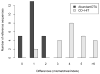
Figure 2
Comparison of the differences between the inferred and known reference sequences. The differences are measured as the total number of mismatchs and indels involved in aligning a reference sequence with the inferred sequence. The difference of 0 means that the inferred sequence is identical to the corresponding reference sequence.
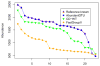
Figure 3
The abundance-rank curves of the Priest09 dataset using different methods. OTUs/clusters are plotted from most to least abundant along the x-axis, with their abundances displayed on the y-axis. The curves only show the high abundant OTUs/clusters. The reference curve shows the best result that any method can achieve, in that the reference sequences are known so that sequencing reads can be mapped to the references directly. The AbundantOTU curve overlaps nicely with the reference curve.
References
- Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009 Sep;6:639–641. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials