Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? - PubMed (original) (raw)
Review
Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)?
Carles Cantó et al. Pharmacol Rev. 2012 Jan.
Abstract
Sirtuin 1 (SIRT1) is an evolutionarily conserved NAD(+)-dependent deacetylase that is at the pinnacle of metabolic control, all the way from yeast to humans. SIRT1 senses changes in intracellular NAD(+) levels, which reflect energy level, and uses this information to adapt the cellular energy output such that it matches cellular energy requirements. The changes induced by SIRT1 activation are generally (but not exclusively) transcriptional in nature and are related to an increase in mitochondrial metabolism and antioxidant protection. These attractive features have validated SIRT1 as a therapeutic target in the management of metabolic disease and prompted an intensive search to identify pharmacological SIRT1 activators. In this review, we first give an overview of the SIRT1 biology with a particular focus on its role in metabolic control. We then analyze the pros and cons of the current strategies used to activate SIRT1 and explore the emerging evidence indicating that modulation of NAD(+) levels could provide an effective way to achieve such goals.
Figures
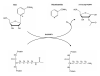
Figure 1. The NAD+-dependent SIRT1 deacetylase reaction
SIRT1 uses NAD+ as a substrate to remove acetyl groups from a target protein. In addition to the deacetylated substrate, the reaction yields nicotinamide and 2′-O-acetyl-ADP ribose as products.
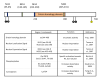
Figure 2. Relevant domains in the human form of the SIRT1 protein
The figure schematizes the span of the conserved sirtuin homology domain as well as the nuclear localization (NLS) and nuclear exportation signals (NES). The residues subject to phosphorylation, by JNK1 and Cyclin/cdk1, and sumoylation are also indicated.
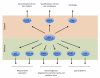
Figure 3. SIRT1 metabolic targets
SIRT1 deacetylates a large arrav of protein targets involved in metabolic regulation. The bottom part of the figure highlights nuclear targets implicated in transcriptional metabolic adaptations. SIRT1’s cytosolic targets are illustrated in the top part. The full names for the abbreviations can be found on the main text.
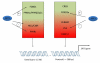
Figure 4. Transcriptional regulation of the SIRT1 gene
Many transcription factors influence the transcriptional activity through acting on both the proximal and distal regions of the SIRT1 promotor. Transcriptional regulators in the green part of the boxes positively regulate SIRT1 gene expression, while those in the red part of the boxes act as negative regulators. Of note, the SIRT1 protein can create many feed-forward loops by deacetylating and enhancing the activity of some positive regulators (FOXOs) while deacetylating and/or inactivating repressor complexes (p53, PPARγ, HIC1/CtBP). Full names for the abbreviations can be found in the text.
Figure 5. Resveratrol promotes mitochondrial biogenesis and lipid oxidation gene expression through indirect AMPK and SIRT1 activation
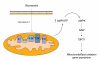
While still a matter of debate, most data currently indicate that the metabolic actions of resveratrol or its metabolites stem from its ability to act as a mild mitochondrial poison, impairing ATP synthesis. The energy stress induced by resveratrol activates AMPK, subsequently stimulating SIRT1 by enhancing NAD+ levels. Then, SIRT1 activates key downstream targets through deacetylation (see Section 1.c), ultimately leading to an adaptative potentiation of mitochondrial biogenesis and lipid oxidation pathways. CI-V represent mitochondrial respiratory complexes I-V. The full names for other abbreviations can be found in the main text.
Figure 6. NAD+ as a nodal point for metabolic regulation
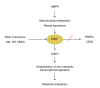
Most evidence to date points out that NAD+ could be rate-limiting for the SIRT1 reaction in diverse conditions. SIRT1 activity would hence be stimulated by interventions which increase NAD+ levels, such as AMPK activation, enhancement of NAD+ biosynthesis through precursor (NA, NR, NMN) supplementation, or through inhibition of alternative NAD+ consuming activities, such as PARPs or CD38. The stimulation of SIRT1 activity by these distinct means improves the capacity of the cells/organism to adapt to external metabolic cues.
Similar articles
- The 2023 Latin America report of the Lancet Countdown on health and climate change: the imperative for health-centred climate-resilient development.
Hartinger SM, Palmeiro-Silva YK, Llerena-Cayo C, Blanco-Villafuerte L, Escobar LE, Diaz A, Sarmiento JH, Lescano AG, Melo O, Rojas-Rueda D, Takahashi B, Callaghan M, Chesini F, Dasgupta S, Posse CG, Gouveia N, Martins de Carvalho A, Miranda-Chacón Z, Mohajeri N, Pantoja C, Robinson EJZ, Salas MF, Santiago R, Sauma E, Santos-Vega M, Scamman D, Sergeeva M, Souza de Camargo T, Sorensen C, Umaña JD, Yglesias-González M, Walawender M, Buss D, Romanello M. Hartinger SM, et al. Lancet Reg Health Am. 2024 Apr 23;33:100746. doi: 10.1016/j.lana.2024.100746. eCollection 2024 May. Lancet Reg Health Am. 2024. PMID: 38800647 Free PMC article. Review. - Exploring conceptual and theoretical frameworks for nurse practitioner education: a scoping review protocol.
Wilson R, Godfrey CM, Sears K, Medves J, Ross-White A, Lambert N. Wilson R, et al. JBI Database System Rev Implement Rep. 2015 Oct;13(10):146-55. doi: 10.11124/jbisrir-2015-2150. JBI Database System Rev Implement Rep. 2015. PMID: 26571290 - Effectiveness of interventions for improving educational outcomes for people with disabilities in low- and middle-income countries: A systematic review.
Hunt X, Saran A, White H, Kuper H. Hunt X, et al. Campbell Syst Rev. 2025 Feb 6;21(1):e70016. doi: 10.1002/cl2.70016. eCollection 2025 Mar. Campbell Syst Rev. 2025. PMID: 39917627 Free PMC article. Review. - Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.
Elwenspoek MM, Thom H, Sheppard AL, Keeney E, O'Donnell R, Jackson J, Roadevin C, Dawson S, Lane D, Stubbs J, Everitt H, Watson JC, Hay AD, Gillett P, Robins G, Jones HE, Mallett S, Whiting PF. Elwenspoek MM, et al. Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
Cited by
- PARP Inhibitors: An Innovative Approach to the Treatment of Inflammation and Metabolic Disorders in Sepsis.
Wasyluk W, Zwolak A. Wasyluk W, et al. J Inflamm Res. 2021 May 6;14:1827-1844. doi: 10.2147/JIR.S300679. eCollection 2021. J Inflamm Res. 2021. PMID: 33986609 Free PMC article. Review. - Intrathecal Injection of SIRT1-modified Human Mesenchymal Stem Cells Alleviates Neuropathic Pain in Rat.
Tian J, Song T, Wang H, Wang W, Zhang Z, Yan R, Ma X, Hu Y. Tian J, et al. J Mol Neurosci. 2021 May;71(5):972-980. doi: 10.1007/s12031-020-01717-2. Epub 2020 Oct 2. J Mol Neurosci. 2021. PMID: 33009636 - SIRT1 enhances glucose tolerance by potentiating brown adipose tissue function.
Boutant M, Joffraud M, Kulkarni SS, García-Casarrubios E, García-Roves PM, Ratajczak J, Fernández-Marcos PJ, Valverde AM, Serrano M, Cantó C. Boutant M, et al. Mol Metab. 2014 Dec 18;4(2):118-31. doi: 10.1016/j.molmet.2014.12.008. eCollection 2015 Feb. Mol Metab. 2014. PMID: 25685699 Free PMC article. - Dihydroxyacetone Exposure Alters NAD(P)H and Induces Mitochondrial Stress and Autophagy in HEK293T Cells.
Smith KR, Hayat F, Andrews JF, Migaud ME, Gassman NR. Smith KR, et al. Chem Res Toxicol. 2019 Aug 19;32(8):1722-1731. doi: 10.1021/acs.chemrestox.9b00230. Epub 2019 Aug 2. Chem Res Toxicol. 2019. PMID: 31328504 Free PMC article. - Role of the AMPK/SIRT1 pathway in non‑alcoholic fatty liver disease (Review).
Anggreini P, Kuncoro H, Sumiwi SA, Levita J. Anggreini P, et al. Mol Med Rep. 2023 Feb;27(2):35. doi: 10.3892/mmr.2022.12922. Epub 2022 Dec 23. Mol Med Rep. 2023. PMID: 36562343 Free PMC article. Review.
References
- Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun. 2006;349(1):353–9. - PubMed
- Altschul R, Hoffer A, Stephen JD. Influence of nicotinic acid on serum cholesterol in man. Arch Biochem Biophys. 1955;54(2):558–9. - PubMed
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274(25):17860–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous