Ribosomal route to small-molecule diversity - PubMed (original) (raw)
. 2012 Jan 11;134(1):418-25.
doi: 10.1021/ja208278k. Epub 2011 Dec 22.
Affiliations
- PMID: 22107593
- PMCID: PMC3268757
- DOI: 10.1021/ja208278k
Ribosomal route to small-molecule diversity
Ma Diarey B Tianero et al. J Am Chem Soc. 2012.
Abstract
The cyanobactin ribosomal peptide (RP) natural product pathway was manipulated to incorporate multiple tandem mutations and non-proteinogenic amino acids, using eight heterologous components simultaneously expressed in Escherichia coli . These studies reveal the potential of RPs for the rational synthesis of complex, new small molecules over multiple-step biosynthetic pathways using simple genetic engineering.
© 2011 American Chemical Society
Figures
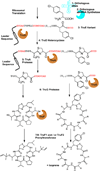
Figure 1. Biosynthesis of tru pathway derivatives in E. coli
Numbered components indicate eight individual elements including enzymes, tRNA, or precursor peptide (TruE variants) that are required for synthesis of mature natural products. The orthologous tRNA and amino acyl tRNA synthetase components required to introduce non-proteinogenic amino acids are shown in blue. Enzymes from the tru pathway are indicated in orange. The precursor peptide TruE is shown with a helical, 35-amino acid leader sequence indicated schematically. X indicates any amino acid.
Figure 2. Expression strategy
Enzymes and two control molecules are synthesized constitutively from the vector ptru-SD1 (top). An internal control sequence and a variable region, that can lead to peptide libraries, is synthesized from ptruE (bottom). Optionally, non-proteinogenic amino acids can be included using a third vector, pEVOL (right).
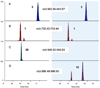
Figure 3. Expression of mutant cyanobactins
E. coli cultures expressing 20 (left) and 15 (right) were harvested and analyzed by FT-MS. Extracted ion chromatograms showed that each culture expressed control compounds 3 (A) and 1 (B). In cells containing plasmids encoding 20, compound 20 could be detected (C), but not 15 (D), nor any other cyanobactin 4–22. Similarly, in cells encoding 15, only compound 15 (D) but not 20 (C), nor any other cyanobactin, could be detected. Thus, each experiment contained two internal positive controls and 19 external negative controls.
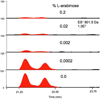
Figure 4. Optimization of non-proteinogenic amino acid incorporation
Compound 10 was synthesized in E. coli and analyzed by LC-ESI-MS. Optimum production was achieved in the absence of inducer (arabinose), whereas higher concentrations of inducer completely repressed synthesis. Two peaks are present because multiple stereoisomers are present (the a-proton adjacent to thiazoline is labile, and Pro undergoes cis-trans isomerization in this family).
Figure 5. Compounds synthesized in this study
Yellow indicates wild-type compounds, while pink bubbles indicate mutations that deviate from wild type. Hexa- (left), hepta- (mid) and octa- (right) peptide derivatives were synthesized using these methods. * indicates compounds for which only mono-prenylated (9, 11, and 12) or non-prenylated (18) derivatives were identified. For all other compounds, both singly, doubly, and sometimes triply prenylated products were identified.
Similar articles
- Comparative anatomy of a regulatory ribosomal protein.
Worbs M, Wahl MC, Lindahl L, Zengel JM. Worbs M, et al. Biochimie. 2002 Aug;84(8):731-43. doi: 10.1016/s0300-9084(02)01410-4. Biochimie. 2002. PMID: 12457561 - A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli.
Ederth J, Mandava CS, Dasgupta S, Sanyal S. Ederth J, et al. Nucleic Acids Res. 2009 Feb;37(2):e15. doi: 10.1093/nar/gkn992. Epub 2008 Dec 11. Nucleic Acids Res. 2009. PMID: 19074194 Free PMC article. - Are ribosomal proteins present at transcription sites on or off ribosomal subunits?
De S, Brogna S. De S, et al. Biochem Soc Trans. 2010 Dec;38(6):1543-7. doi: 10.1042/BST0381543. Biochem Soc Trans. 2010. PMID: 21118123 Review. - Ribosome engineering and secondary metabolite production.
Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K. Ochi K, et al. Adv Appl Microbiol. 2004;56:155-84. doi: 10.1016/S0065-2164(04)56005-7. Adv Appl Microbiol. 2004. PMID: 15566979 Review. No abstract available.
Cited by
- Efficient genetic code expansion without host genome modifications.
Costello A, Peterson AA, Lanster DL, Li Z, Carver GD, Badran AH. Costello A, et al. Nat Biotechnol. 2024 Sep 11. doi: 10.1038/s41587-024-02385-y. Online ahead of print. Nat Biotechnol. 2024. PMID: 39261591 - Cellular Site-Specific Incorporation of Noncanonical Amino Acids in Synthetic Biology.
Niu W, Guo J. Niu W, et al. Chem Rev. 2024 Sep 25;124(18):10577-10617. doi: 10.1021/acs.chemrev.3c00938. Epub 2024 Aug 29. Chem Rev. 2024. PMID: 39207844 Review. - An Autocatalytic Peptide Cyclase Improves Fidelity and Yield of Circular Peptides In Vivo and In Vitro.
Lacerna N 2nd, Cong Y, Schmidt EW. Lacerna N 2nd, et al. ACS Synth Biol. 2024 Jan 19;13(1):394-401. doi: 10.1021/acssynbio.3c00645. Epub 2024 Jan 9. ACS Synth Biol. 2024. PMID: 38194299 Free PMC article. - Incorporation of Non-Canonical Amino Acids into Antimicrobial Peptides: Advances, Challenges, and Perspectives.
Du Y, Li L, Zheng Y, Liu J, Gong J, Qiu Z, Li Y, Qiao J, Huo YX. Du Y, et al. Appl Environ Microbiol. 2022 Dec 13;88(23):e0161722. doi: 10.1128/aem.01617-22. Epub 2022 Nov 23. Appl Environ Microbiol. 2022. PMID: 36416555 Free PMC article. Review. - De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
Vinogradov AA, Zhang Y, Hamada K, Chang JS, Okada C, Nishimura H, Terasaka N, Goto Y, Ogata K, Sengoku T, Onaka H, Suga H. Vinogradov AA, et al. J Am Chem Soc. 2022 Nov 9;144(44):20332-20341. doi: 10.1021/jacs.2c07937. Epub 2022 Oct 25. J Am Chem Soc. 2022. PMID: 36282922 Free PMC article.
References
- Mitchell W. Curr. Opin. Chem. Biol. 2011;15:505. - PubMed
- Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM071425/GM/NIGMS NIH HHS/United States
- GM097206/GM/NIGMS NIH HHS/United States
- R01 GM071425-05/GM/NIGMS NIH HHS/United States
- GM071425-S1/GM/NIGMS NIH HHS/United States
- R01 GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-01A1/GM/NIGMS NIH HHS/United States
- R01 GM071425-04/GM/NIGMS NIH HHS/United States
- R01 GM071425-03/GM/NIGMS NIH HHS/United States
- R01 GM097206/GM/NIGMS NIH HHS/United States
- R01 GM071425-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources