Phytozome: a comparative platform for green plant genomics - PubMed (original) (raw)
. 2012 Jan;40(Database issue):D1178-86.
doi: 10.1093/nar/gkr944. Epub 2011 Nov 22.
Affiliations
- PMID: 22110026
- PMCID: PMC3245001
- DOI: 10.1093/nar/gkr944
Phytozome: a comparative platform for green plant genomics
David M Goodstein et al. Nucleic Acids Res. 2012 Jan.
Abstract
The number of sequenced plant genomes and associated genomic resources is growing rapidly with the advent of both an increased focus on plant genomics from funding agencies, and the application of inexpensive next generation sequencing. To interact with this increasing body of data, we have developed Phytozome (http://www.phytozome.net), a comparative hub for plant genome and gene family data and analysis. Phytozome provides a view of the evolutionary history of every plant gene at the level of sequence, gene structure, gene family and genome organization, while at the same time providing access to the sequences and functional annotations of a growing number (currently 25) of complete plant genomes, including all the land plants and selected algae sequenced at the Joint Genome Institute, as well as selected species sequenced elsewhere. Through a comprehensive plant genome database and web portal, these data and analyses are available to the broader plant science research community, providing powerful comparative genomics tools that help to link model systems with other plants of economic and ecological importance.
Figures
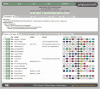
Figure 1.
Default view of the Gene Family page for a 17 member core eudicot family. Members are listed according to their order in the tree on the Phytozome home page. The membership class of each gene is indicated in the leftmost column (
Supplementary Methods S2
). For each member, Gene Page and GBrowse links are provided, as well as links to external databases (if these exist), aliases, symbols and deflines. The synteny view in the right column shows the five upstream and five downstream neighbors of each family member (who are rendered as gray icons in the middle of each synteny row). Each syntenic segment is oriented to render family members in the same orientation (consistent with their presumed descent from a common ancestor). Gene icons sharing the same (non-white) color are all members of the same gene family at this node; this can provide syntenic support for the hypothesis of a common ancestor for family members.
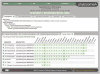
Figure 2.
Family History view of the gene family in Figure 1. All the descendants and ancestors of this core eudicot family (which is highlighted) are visible in the history view. The strict nesting of families is observed, though one needs to remember that one of the Eucalyptus genes in this core eudicot family is an incomplete pledge (
Supplementary Methods S2
), and is not present in the deeper Embryophyte and Viridiplantae ancestors.
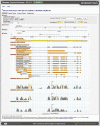
Figure 3.
GBrowse view of the local genomic context of the poplar gene from the family in Figure 1. Primary and alternative transcripts (if present), assembled EST data and related plant peptides are shown aligned against the genome. Not shown are tracks of repetitive regions, GC content and the alignment of ESTs from related species. Interspecies whole genome alignments, displayed in the VISTA tracks, reveal the tendency towards strong genomic sequence conservation in coding regions (which are under selective pressure), which weakens as one considers more distantly related species (e.g. rice-poplar versus the more closely related eucalyptus-poplar VISTA alignments). Displayed gene models are hyperlinked to their respective gene pages.
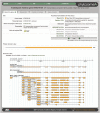
Figure 4.
Default view of the Gene Page for the Arabidopsis thaliana gene in the family of Figure 1, showing primary transcript info, functional annotations and simplified genomic context. This locus has an alternative transcript (which appears to differ primarily in its 5′-UTR). Note the strong splicing support provided by the BLATX aligned Arabidopsis lyrata peptide (which in actuality is also a member of this family).
Similar articles
- Closing target trimming and CTTdocker programs for discovering hidden superfamily loci in genomes.
Hua Z, Early MJ. Hua Z, et al. PLoS One. 2019 Jul 2;14(7):e0209468. doi: 10.1371/journal.pone.0209468. eCollection 2019. PLoS One. 2019. PMID: 31265455 Free PMC article. - PLAZA: a comparative genomics resource to study gene and genome evolution in plants.
Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. Proost S, et al. Plant Cell. 2009 Dec;21(12):3718-31. doi: 10.1105/tpc.109.071506. Epub 2009 Dec 29. Plant Cell. 2009. PMID: 20040540 Free PMC article. - Gramene 2013: comparative plant genomics resources.
Monaco MK, Stein J, Naithani S, Wei S, Dharmawardhana P, Kumari S, Amarasinghe V, Youens-Clark K, Thomason J, Preece J, Pasternak S, Olson A, Jiao Y, Lu Z, Bolser D, Kerhornou A, Staines D, Walts B, Wu G, D'Eustachio P, Haw R, Croft D, Kersey PJ, Stein L, Jaiswal P, Ware D. Monaco MK, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D1193-9. doi: 10.1093/nar/gkt1110. Epub 2013 Nov 11. Nucleic Acids Res. 2014. PMID: 24217918 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - The Chlamydomonas genome project: a decade on.
Blaby IK, Blaby-Haas CE, Tourasse N, Hom EF, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, King S, Witman GB, Stanke M, Harris EH, Goodstein D, Grimwood J, Schmutz J, Vallon O, Merchant SS, Prochnik S. Blaby IK, et al. Trends Plant Sci. 2014 Oct;19(10):672-80. doi: 10.1016/j.tplants.2014.05.008. Epub 2014 Jun 17. Trends Plant Sci. 2014. PMID: 24950814 Free PMC article. Review.
Cited by
- Genome-Wide Identification of the Remorin Gene Family in Poplar and Their Responses to Abiotic Stresses.
Li Z, Wang H, Li C, Liu H, Luo J. Li Z, et al. Life (Basel). 2024 Sep 27;14(10):1239. doi: 10.3390/life14101239. Life (Basel). 2024. PMID: 39459539 Free PMC article. - Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds.
Song JH, Shin G, Kim HJ, Lee SB, Moon JY, Jeong JC, Choi HK, Kim IA, Song HJ, Kim CY, Chung YS. Song JH, et al. Int J Mol Sci. 2022 Sep 13;23(18):10583. doi: 10.3390/ijms231810583. Int J Mol Sci. 2022. PMID: 36142495 Free PMC article. - Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon.
Mandadi KK, Scholthof KB. Mandadi KK, et al. Plant Cell. 2015 Jan;27(1):71-85. doi: 10.1105/tpc.114.133991. Epub 2015 Jan 29. Plant Cell. 2015. PMID: 25634987 Free PMC article. - Mannans and endo-β-mannanases (MAN) in Brachypodium distachyon: expression profiling and possible role of the BdMAN genes during coleorhiza-limited seed germination.
González-Calle V, Barrero-Sicilia C, Carbonero P, Iglesias-Fernández R. González-Calle V, et al. J Exp Bot. 2015 Jul;66(13):3753-64. doi: 10.1093/jxb/erv168. Epub 2015 Apr 28. J Exp Bot. 2015. PMID: 25922488 Free PMC article. - Establishing Chlamydomonas reinhardtii as an industrial biotechnology host.
Scaife MA, Nguyen GTDT, Rico J, Lambert D, Helliwell KE, Smith AG. Scaife MA, et al. Plant J. 2015 May;82(3):532-546. doi: 10.1111/tpj.12781. Epub 2015 Mar 8. Plant J. 2015. PMID: 25641561 Free PMC article. Review.
References
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The sol genomics network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources