Gene expression databases for kidney epithelial cells - PubMed (original) (raw)
Review
Gene expression databases for kidney epithelial cells
Jennifer C Huling et al. Am J Physiol Renal Physiol. 2012.
Abstract
The 21st century has seen an explosion of new high-throughput data from transcriptomic and proteomic studies. These data are highly relevant to the design and interpretation of modern physiological studies but are not always readily accessible to potential users in user-friendly, searchable formats. Data from our own studies involving transcriptomic and proteomic profiling of renal tubule epithelia have been made available on a variety of online databases. Here, we provide a roadmap to these databases and illustrate how they may be useful in the design and interpretation of physiological studies. The databases can be accessed through http://helixweb.nih.gov/ESBL/Database.
Figures
Fig. 1.
The transcriptomic database webpage presents the user with 2 options for accessing information: links to each of the 3 segment-specific databases individually by clicking on the appropriate database title (A) and a Basic Local Alignment Search Tool (BLAST) search of all 3 databases at once by inputting the amino acid sequence into the input box (B). The “Results” page will display all similar sequences from the transcriptomic databases and indicate which databases contain each matched transcript. (Note that to avoid scrolling, it may be necessary to increase screen resolution for this page and subsequent pages.)
Fig. 2.
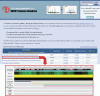
Sample output of a BLAST search. The Results page gives a brief explanation, a diagram of the nephron for reference, and a summary table displaying the key results of the search. The table lists matches based on amino acid sequence and for each protein displays general information about the protein and about the protein's location. A: 3 columns highlighted correspond to each of the 3 segment-specific databases being searched. A green icon in a column means that the protein does appear in the corresponding database, and a red icon means the protein does not (determined if the Probe Set ID had a signal strength <0.2). The numbers beside each icon are the signal strengths. B: when the user holds the cursor over the word “view” a visual summary of the previous three columns is shown. The signal strengths are displayed over the correct nephron region. Each region is also shaded in blue, with darker blue shading corresponding to a higher signal level. IMCD, inner medullary collecting duct.
Fig. 3.
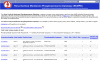
Transcriptomic database for the medullary thick ascending limb (TAL). The page provides a description of the methods used to collect the data and displays information for all of the Affymetrix Probe Set IDs found in the TAL. Each probe set ID is listed along with the associated protein name, gene symbol, accession number, Swiss-Prot number, and the fluorescent signal strength in the array readout. The database also includes the following features: the option to sort the database (A), a link to the BLAST search featured on the main page so that users can find a specific protein by amino acid sequence (B), a link that downloads the data into a spreadsheet (C), the Swiss-Prot function of the proteins visible by holding the cursor over the Swiss-Prot number (D).
Fig. 4.
IMCD Proteome Database contains the same features as the transcriptome databases (sorting options, BLAST search, downloadable data, and mouse-over Swiss-Prot functions). In addition, the name of each protein links the user to the “Protein Viewer,” which displays various features mapped along the length of the protein's amino acid sequence. The “Tech view” feature allows the user to find the correct reference with the appropriate description of the techniques used to generate the data.
Fig. 5.
Phosphoproteomic database for proximal tubule and other renal cortical membrane proteins. The page provides a sortable table of phosphorylation sites detected along with the accession number, official gene symbol, protein name, phosphopeptide sequence detected, and site assignment information. In addition the user is able to download all the data as a spreadsheet or a PDF file.
Similar articles
- Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes.
Lee JW, Chou CL, Knepper MA. Lee JW, et al. J Am Soc Nephrol. 2015 Nov;26(11):2669-77. doi: 10.1681/ASN.2014111067. Epub 2015 Mar 27. J Am Soc Nephrol. 2015. PMID: 25817355 Free PMC article. - Quantitative Proteomics of All 14 Renal Tubule Segments in Rat.
Limbutara K, Chou CL, Knepper MA. Limbutara K, et al. J Am Soc Nephrol. 2020 Jun;31(6):1255-1266. doi: 10.1681/ASN.2020010071. Epub 2020 May 1. J Am Soc Nephrol. 2020. PMID: 32358040 Free PMC article. - BIG: a large-scale data integration tool for renal physiology.
Zhao Y, Yang CR, Raghuram V, Parulekar J, Knepper MA. Zhao Y, et al. Am J Physiol Renal Physiol. 2016 Oct 1;311(4):F787-F792. doi: 10.1152/ajprenal.00249.2016. Epub 2016 Jun 8. Am J Physiol Renal Physiol. 2016. PMID: 27279488 Free PMC article. - Transport and metabolic functions in cultured renal tubule cells.
Horster MF, Stopp M. Horster MF, et al. Kidney Int. 1986 Jan;29(1):46-53. doi: 10.1038/ki.1986.7. Kidney Int. 1986. PMID: 3515014 Review. - Sodium entry pathways in renal epithelial cell lines.
Saier MH Jr, Boerner P, Grenier FC, McRoberts JA, Rindler MJ, Taub M, U HS. Saier MH Jr, et al. Miner Electrolyte Metab. 1986;12(1):42-50. Miner Electrolyte Metab. 1986. PMID: 2421147 Review.
Cited by
- Prostaglandin E2 EP3 receptor regulates cyclooxygenase-2 expression in the kidney.
Vio CP, Quiroz-Munoz M, Cuevas CA, Cespedes C, Ferreri NR. Vio CP, et al. Am J Physiol Renal Physiol. 2012 Aug 1;303(3):F449-57. doi: 10.1152/ajprenal.00634.2011. Epub 2012 May 23. Am J Physiol Renal Physiol. 2012. PMID: 22622465 Free PMC article. - Drug-Like Protein-Protein Interaction Modulators: Challenges and Opportunities for Drug Discovery and Chemical Biology.
Villoutreix BO, Kuenemann MA, Poyet JL, Bruzzoni-Giovanelli H, Labbé C, Lagorce D, Sperandio O, Miteva MA. Villoutreix BO, et al. Mol Inform. 2014 Jun;33(6-7):414-437. doi: 10.1002/minf.201400040. Epub 2014 Jun 2. Mol Inform. 2014. PMID: 25254076 Free PMC article. Review. - Application of systems biology principles to protein biomarker discovery: urinary exosomal proteome in renal transplantation.
Pisitkun T, Gandolfo MT, Das S, Knepper MA, Bagnasco SM. Pisitkun T, et al. Proteomics Clin Appl. 2012 Jun;6(5-6):268-78. doi: 10.1002/prca.201100108. Proteomics Clin Appl. 2012. PMID: 22641613 Free PMC article. - Identification of Inflammatory Proteomics Networks of Toll-like Receptor 4 through Immunoprecipitation-Based Chemical Cross-Linking Proteomics.
Shahinuzzaman ADA, Kamal AHM, Chakrabarty JK, Rahman A, Chowdhury SM. Shahinuzzaman ADA, et al. Proteomes. 2022 Sep 1;10(3):31. doi: 10.3390/proteomes10030031. Proteomes. 2022. PMID: 36136309 Free PMC article. - Lack of an effect of nephron-specific deletion of adenylyl cyclase 3 on renal sodium and water excretion or arterial pressure.
Kittikulsuth W, Stuart D, Van Hoek AN, Kohan DE. Kittikulsuth W, et al. Physiol Rep. 2015 Mar;3(3):e12316. doi: 10.14814/phy2.12316. Physiol Rep. 2015. PMID: 25747587 Free PMC article.
References
- Chabardes-Garonne D, Mejean A, Aude JC, Cheval L, Di SA, Gaillard MC, Imbert-Teboul M, Wittner M, Balian C, Anthouard V, Robert C, Segurens B, Wincker P, Weissenbach J, Doucet A, Elalouf JM. A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci USA 100: 13710–13715, 2003 - PMC - PubMed
- Cheval L, Morla L, Elalouf JM, Doucet A. Kidney collecting duct acid-base “regulon.” Physiol Genomics 27: 271–281, 2006 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources