RING domain mutations uncouple TRIM5α restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating - PubMed (original) (raw)
RING domain mutations uncouple TRIM5α restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating
Amanda Roa et al. J Virol. 2012 Feb.
Abstract
Rhesus TRIM5α (TRIM5α(rh)) is a cytosolic protein that potently restricts HIV-1 at an early postentry stage, prior to reverse transcription. The ability of TRIM5α(rh) to block HIV-1 infection has been correlated with a decrease of pelletable HIV-1 capsid during infection. To genetically dissect the ability of TRIM5α to block reverse transcription, we studied a set of TRIM5α(rh) RING domain mutants that potently restrict HIV-1 but allow the occurrence of reverse transcription. These TRIM5α(rh) RING variants blocked HIV-1 infection after reverse transcription but prior to integration, as suggested by the routing of nuclear viral DNA to circularization in the form of 2-long terminal repeat (2-LTR) circles. The folding of RING domain variants was similar to that of the wild type, as evaluated by nuclear magnetic resonance. RING domain changes that allowed the occurrence of reverse transcription were impaired in their ability to decrease the amount of pelletable capsid compared with wild-type TRIM5α. Similar effects of this particular group of mutations were observed with human TRIM5α inhibition of N-tropic murine leukemia virus (N-MLV). Interestingly, TRIM5α(rh) RING domain variants also prevented the degradation of TRIM5α(rh) that occurs following cell entry of HIV-1. These data correlated the block of reverse transcription with the ability of TRIM5α to accelerate uncoating. Collectively, these results suggest that TRIM5α(rh) blocks HIV-1 reverse transcription by inducing premature viral uncoating in target cells.
Figures
Fig 1
Restriction of HIV-1 and N-MLV infection after reverse transcription. Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5α proteins. Stable cell lines were selected with 5 μg/ml of puromycin. CF2Th cell lines expressing the indicated wild-type and TRIM5α RING domain variants or containing the empty LPCX vector were challenged with different amounts of HIV-1–GFP (A) or N-MLV–GFP (C). Infection was determined by measuring the number of GFP-positive cells 48 h postinfection by flow cytometry. Similar results were obtained in three independent experiments, and the result of one experiment is shown. Similarly, CF2Th cell lines expressing the indicated wild-type and mutant TRIM5α proteins or containing the empty LPCX vector were challenged at an MOI of 0.4 with DNase-pretreated HIV-1–GFP (B) or N-MLV–GFP (D) viruses. After 7 h, cells were lysed and total DNA was extracted. The levels of viral DNA were measured by quantitative real-time PCR, using a probe against GFP, as described in Materials and Methods. Similar results were obtained in three independent experiments, and the standard deviation is shown. (E) The expression level of mutant and wild-type TRIM5α proteins was assayed by Western blotting using HRP-conjugated antibodies against HA. (F) CF2Th cell lines expressing the indicated wild-type and mutant TRIM5αrh proteins or containing the empty LPCX vector were challenged at an MOI of 0.4 with DNase-pretreated VSV-G-pseudotyped Env-defective HIV-1 viruses (R9 clone). After 24 h, cells were lysed and total DNA was extracted. The levels of HIV-1 2-LTR circles were measured by quantitative real-time PCR, using specific primers to detect the junction of 2-LTR circles, as described in Materials and Methods. Similar results were obtained in three independent experiments, and the standard deviation is shown. In panels A and C, black arrows point out the viral doses of HIV-1 and N-MLV used to measure reverse transcription.
Fig 2
Protein folding of RING domain mutants. (A) Structure of the RING domain of TRIM5αrh illustrating the position of tyrosine 63 (red). The side chain of tyrosine 63 is part of the hydrophobic core of the RING domain and is surrounded by the hydrophobic side chains of residues L28, I39, I61, P65, and I68. (B) The two-dimensional 1H, 15NHSQC spectra of wild-type (black) and Y63E (red) RING domains are shown. The overlay of the spectral data (black and red) showed that the RING domain Y63E mutation does not disrupt protein folding.
Fig 3
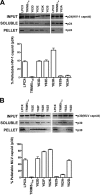
Fate of the retroviral capsid in cells expressing TRIM5α variants. (A) CF2Th cell lines expressing the indicated wild-type and mutant TRIM5αrh proteins or containing the empty LPCX vector were incubated with equivalent amounts of HIV-1–GFP at 4°C for 30 min. The cells were washed and returned to 37°C, and infection was allowed to proceed for 12 h. Cell extracts were fractionated on sucrose gradients as described in Materials and Methods. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against the HIV-1 p24 capsid protein. (B) CF2Th cell lines expressing the indicated wild-type and mutant TRIM5αhu proteins or containing the empty LPCX vector were incubated with similar amounts of N-MLV–GFP at 4°C for 30 min. The cells were washed and returned to 37°C, and infection was allowed to proceed for 4 h. Cell extracts were fractionated on sucrose gradients as described in Materials and Methods. Input, soluble, and pellet fractions were analyzed by Western blotting using antibodies against the N-MLV p30 capsid protein. The lower bar graphs represent the fluorescent quantification of pelletable capsid shown as a percentage. Similar results were obtained in three independent experiments and the standard deviation is shown.
Fig 4
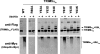
E3 ubiquitin ligase activity of TRIM5αrh RING domain variants. Human 293T cells were transfected with plasmids encoding FLAG-tagged mutant and wild-type TRIM5αrh proteins. Forty-eight hours later, the cells expressing each TRIM5αrh variant were lysed in whole-cell extract and immunoprecipitated using anti-FLAG-agarose beads as described in Materials and Methods. Beads containing the immunoprecipitated TRIM5αrh variants were washed and eluted with 200 μg/ml of FLAG tripeptide in whole-cell extract buffer as described in Materials and Methods. Samples were supplemented with 5 μM ubiquitin aldehyde, a potent inhibitor of all ubiquitin C-terminal hydrolases, ubiquitin-specific proteases, and deubiquitinating enzymes. Similar amounts of inhibitor-treated samples containing mutant and wild-type TRIM5αrh were incubated with 200 nM enzyme E1 (human recombinant UBE1), 200 μM ubiquitin tagged with a myc epitope (human recombinant ubiquitin), and an energy regeneration solution containing MgCl2, ATP, and ATP-regenerating enzymes to recycle hydrolyzed ATP. The final reaction was supplemented or not with 100 nM enzyme E2 (human recombinant UbcH5a) as indicated. The reaction mixture was incubated at 37°C for 1 h, and collected fractions were analyzed by Western blotting using HRP-conjugated antibodies against FLAG to detect the levels of TRIM5αrh variants (anti-FLAG). To detect TRIM5αrh ubiquitylated forms, membranes were blotted using HRP-conjugated antibodies against myc (anti-myc). The results of three independent experiments were similar; the result of a single experiment is shown.
Fig 5
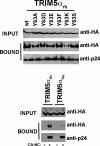
Binding of TRIM5αrh RING domain variants to the HIV-1 CA. 293T cells were transfected with plasmids expressing the indicated wild-type and mutant TRIM5αrh proteins tagged with HA epitopes. Thirty-six hours after transfection, cells were lysed. The lysates were incubated at room temperature for 1 h with HIV-1 CA-NC complexes that had been assembled in vitro. The mixtures were applied onto a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted using anti-HA antibodies. The pellet from the 70% cushion (BOUND) was analyzed by Western blotting using antibodies against the HA tag and HIV-1 CA-NC protein. The blots were quantitated as described in Materials and Methods, and values for binding are shown in Tables 1 and 2. The results of three independent experiments were similar; the result of a single experiment is shown.
Fig 6
TRIM5αrh RING domain variant higher-order self-association (HOSA). 293T cells were transfected with plasmids expressing the indicated wild-type or mutant TRIM5α proteins with a FLAG or HA epitope tag. Cells expressing wild-type and mutant TRIM5αrh proteins were lysed 48 h after transfection. The cell lysates containing similar inputs were mixed (INPUT), and the indicated mixtures were used for immunoprecipitation by an antibody directed against the FLAG epitope, as described in Materials and Methods. Elution of the immunocomplexes was performed with a FLAG tripeptide and analyzed by Western blotting using anti-HA and anti-FLAG antibodies (higher-order self-associates). The results of three independent experiments were similar; the result of a single experiment is shown.
Fig 7
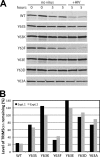
HIV-1-induced degradation of TRIM5αrh RING domain variants. (A) TRIM5αrh-expressing Cf2Th cell lines were cultured in cycloheximide-containing media in the presence or absence of HIV-1 particles for the indicated time periods, and cytoplasmic lysates were prepared and analyzed by immunoblotting for TRIM5αrh. Due to the lower steady-state levels of the wild-type and Y63F mutant TRIM5αrh proteins, the panels for these samples were obtained by scanning the immunoblot with the LI-COR Odyssey at a higher laser intensity than for the other mutants. (B) Quantification of the effect of HIV-1 on TRIM5αrh protein levels after 5 h of culture. Values shown are the averages of duplicate parallel assays. Paired values depict the results from two independent experiments.
Similar articles
- Virus-specific effects of TRIM5α(rh) RING domain functions on restriction of retroviruses.
Li X, Kim J, Song B, Finzi A, Pacheco B, Sodroski J. Li X, et al. J Virol. 2013 Jul;87(13):7234-45. doi: 10.1128/JVI.00620-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637418 Free PMC article. - Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus.
Kim J, Tipper C, Sodroski J. Kim J, et al. J Virol. 2011 Aug;85(16):8116-32. doi: 10.1128/JVI.00341-11. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680520 Free PMC article. - The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid.
Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. Perron MJ, et al. J Virol. 2007 Mar;81(5):2138-48. doi: 10.1128/JVI.02318-06. Epub 2006 Nov 29. J Virol. 2007. PMID: 17135314 Free PMC article. - Retroviral restriction factors TRIM5α: therapeutic strategy to inhibit HIV-1 replication.
Zhang J, Ge W, Zhan P, De Clercq E, Liu X. Zhang J, et al. Curr Med Chem. 2011;18(17):2649-54. doi: 10.2174/092986711795933687. Curr Med Chem. 2011. PMID: 21568899 Review. - Recent insights into the mechanism and consequences of TRIM5α retroviral restriction.
Sastri J, Campbell EM. Sastri J, et al. AIDS Res Hum Retroviruses. 2011 Mar;27(3):231-8. doi: 10.1089/AID.2010.0367. AIDS Res Hum Retroviruses. 2011. PMID: 21247355 Free PMC article. Review.
Cited by
- Retrovirus restriction by TRIM5 proteins requires recognition of only a small fraction of viral capsid subunits.
Shi J, Friedman DB, Aiken C. Shi J, et al. J Virol. 2013 Aug;87(16):9271-8. doi: 10.1128/JVI.00713-13. Epub 2013 Jun 19. J Virol. 2013. PMID: 23785198 Free PMC article. - Potent restriction of HIV-1 and SIVmac239 replication by African green monkey TRIM5α.
Coren LV, Trivett MT, Jain S, Ayala VI, Del Prete GQ, Ohlen C, Ott DE. Coren LV, et al. Retrovirology. 2015 Feb 7;12:11. doi: 10.1186/s12977-015-0137-9. Retrovirology. 2015. PMID: 25809491 Free PMC article. - A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5α and mammalian autophagy-related ATG8 proteins.
Keown JR, Black MM, Ferron A, Yap M, Barnett MJ, Pearce FG, Stoye JP, Goldstone DC. Keown JR, et al. J Biol Chem. 2018 Nov 23;293(47):18378-18386. doi: 10.1074/jbc.RA118.004202. Epub 2018 Oct 3. J Biol Chem. 2018. PMID: 30282803 Free PMC article. - HIV-1 uncoating: connection to nuclear entry and regulation by host proteins.
Ambrose Z, Aiken C. Ambrose Z, et al. Virology. 2014 Apr;454-455:371-9. doi: 10.1016/j.virol.2014.02.004. Epub 2014 Feb 20. Virology. 2014. PMID: 24559861 Free PMC article. Review. - Restriction of HIV-1 and other retroviruses by TRIM5.
Ganser-Pornillos BK, Pornillos O. Ganser-Pornillos BK, et al. Nat Rev Microbiol. 2019 Sep;17(9):546-556. doi: 10.1038/s41579-019-0225-2. Epub 2019 Jul 16. Nat Rev Microbiol. 2019. PMID: 31312031 Free PMC article. Review.
References
- Aoki M, et al. 2009. Automated system for high-throughput protein production using the dialysis cell-free method. Protein Expr. Purif. 68: 128–136 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 MH086162/MH/NIMH NIH HHS/United States
- R01 AI076121/AI/NIAID NIH HHS/United States
- R01 AI087390/AI/NIAID NIH HHS/United States
- 4R00MH086162-02/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources