Comparative genomics of the Type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins - PubMed (original) (raw)
Comparative genomics of the Type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins
Pieter De Maayer et al. BMC Genomics. 2011.
Abstract
Background: The Type VI secretion apparatus is assembled by a conserved set of proteins encoded within a distinct locus. The putative effector proteins Hcp and VgrG are also encoded within these loci. We have identified numerous distinct Type VI secretion system (T6SS) loci in the genomes of several ecologically diverse Pantoea and Erwinia species and detected the presence of putative effector islands associated with the hcp and vgrG genes.
Results: Between two and four T6SS loci occur among the Pantoea and Erwinia species. While two of the loci (T6SS-1 and T6SS-2) are well conserved among the various strains, the third (T6SS-3) locus is not universally distributed. Additional orthologous loci are present in Pantoea sp. aB-valens and Erwinia billingiae Eb661. Comparative analysis of the T6SS-1 and T6SS-3 loci showed non-conserved islands associated with the vgrG and hcp, and vgrG genes, respectively. These regions had a G+C content far lower than the conserved portions of the loci. Many of the proteins encoded within the hcp and vgrG islands carry conserved domains, which suggests they may serve as effector proteins for the T6SS. A number of the proteins also show homology to the C-terminal extensions of evolved VgrG proteins.
Conclusions: Extensive diversity was observed in the number and content of the T6SS loci among the Pantoea and Erwinia species. Genomic islands could be observed within some of T6SS loci, which are associated with the hcp and vgrG proteins and carry putative effector domain proteins. We propose new hypotheses concerning a role for these islands in the acquisition of T6SS effectors and the development of novel evolved VgrG and Hcp proteins.
Figures
Figure 1
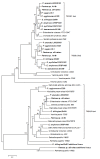
Distribution of T6SS loci among Pantoea and Erwinia species. The presence (+) or absence (-) of ortholgous T6SS loci for the various Pantoea and Erwinia strains are shown. A neighbor-joining tree (bootstrap n = 1,000) based on a ClustalW alignment of GyrB amino acid sequences indicates the phylogenetic relationship of the strains.
Figure 2
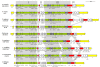
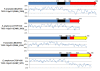
The T6SS-1 loci of P. ananatis LMG 20103 and E. amylovora CFBP 1430. The conserved regions (block I and III) are shaded in gray, while the non-conserved hcp and vgrG islands are not shaded. Genes encoding conserved domain proteins are represented by green arrows while the grey arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-1 loci which were not identified as part of the conserved core described by Boyer et al. [15]. Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white. Graphs show the G+C content (%) (window size = 50 bp, step = 10 bp) in the respective T6SS-1 loci.
Figure 3
The orthologous T6SS-1 loci in Pantoea and Erwinia species. The conserved regions (block I and III) are shaded in gray, while the non-conserved hcp and vgrG islands are not shaded. Genes encoding conserved domain proteins identified by Boyer et al. [15] are represented by green arrows, and grey arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-1 loci which were not identified as part of the conserved core described by Boyer et al. [15]. Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white.
Figure 4
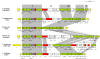
The orthologous T6SS-2 loci in Pantoea and Erwinia species. Genes encoding proteins with the conserved domains identified by Boyer et al. [15] are represented by green arrows while the grey arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-2 loci which were not identified as part of the conserved core. White arrows represent the genes not conserved among the Pantoea and Erwinia species.
Figure 5
Phylogenetic relationships between the T6SS loci among the Pantoea and Erwinia species. A neighbor-joining tree (bootstrap n = 1,000; Poisson correction; Complete gap deletion) was constructed based on a ClustalW alignment of the IcmF (COG3523) amino acid sequences from the Pantoea and Erwinia T6SS loci and orthologous loci in several closely related Enterobacteriaceae.
Figure 6
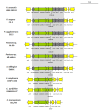
The T6SS-3 loci of P. ananatis LMG 20103 and E. amylovora CFBP 1430. The conserved regions (block I, III and V) are shaded in gray, while non-conserved regions are not shaded. Genes encoding conserved domain proteins identified by Boyer et al. [15] represented by green arrows while the grey arrow indicates a gene conserved among the Pantoea and Erwinia T6SS-3 loci which was not identified as part of the conserved core. Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white. The graphs show the G+C content (%) (window size = 50 bp, step = 10 bp) in the respective T6SS-3 loci.
Figure 7
The orthologous T6SS-3 loci in Pantoea and Erwinia species. The conserved regions (block I, III and V) are shaded in gray, while non-conserved regions are not shaded. Genes encoding conserved core proteins identified by Boyer et al. [15] are represented by green arrows while the grey arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-3 loci which were not identified as part of the conserved core. Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white.
Figure 8
Additional T6SS loci in Pantoea sp. aB-valens and Erwinia billingiae Eb661. Genes encoding proteins with the conserved domains identified by Boyer et al. [15] are indicated by green arrows. Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white.
Figure 9
Conserved and non-conserved domains in the VgrG proteins of P. ananatis LMG 20103 and E. amylovora CFBP 1430. The conserved VgrG domains are colored in blue, Gp5 domains are in black, COG4253 domains in the T6SS-3 VgrG proteins in yellow and the non-conserved C-terminal extensions are colored in red. The amino acid locations of each domain are shown. The graphs show G+C contents (window size = 50 bp, step = 10 bp) in the respective vgrG genes.
Figure 10
Phylogeny of the Pantoea and Erwinia VgrG proteins. Neighbor-joining tree showing the phylogenetic relationship between the VgrG proteins among the various Pantoea and Erwinia species. The tree (bootstrap n = 1,000; Poisson correction; Complete gap deletion) was constructed based on a ClustalW alignment of the VgrG amino acid sequences. "Evolved" VgrG proteins are indicated in brackets.
Similar articles
- Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes.
Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Blondel CJ, et al. BMC Genomics. 2009 Aug 4;10:354. doi: 10.1186/1471-2164-10-354. BMC Genomics. 2009. PMID: 19653904 Free PMC article. - Comparative genomics of type VI secretion systems in strains of Pantoea ananatis from different environments.
Shyntum DY, Venter SN, Moleleki LN, Toth I, Coutinho TA. Shyntum DY, et al. BMC Genomics. 2014 Feb 26;15:163. doi: 10.1186/1471-2164-15-163. BMC Genomics. 2014. PMID: 24571088 Free PMC article. - Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis.
Sha J, Rosenzweig JA, Kozlova EV, Wang S, Erova TE, Kirtley ML, van Lier CJ, Chopra AK. Sha J, et al. Microbiology (Reading). 2013 Jun;159(Pt 6):1120-1135. doi: 10.1099/mic.0.063495-0. Epub 2013 Mar 21. Microbiology (Reading). 2013. PMID: 23519162 Free PMC article. - The bacterial type VI secretion machine: yet another player for protein transport across membranes.
Filloux A, Hachani A, Bleves S. Filloux A, et al. Microbiology (Reading). 2008 Jun;154(Pt 6):1570-1583. doi: 10.1099/mic.0.2008/016840-0. Microbiology (Reading). 2008. PMID: 18524912 Review. - Tubules and donuts: a type VI secretion story.
Bönemann G, Pietrosiuk A, Mogk A. Bönemann G, et al. Mol Microbiol. 2010 May;76(4):815-21. doi: 10.1111/j.1365-2958.2010.07171.x. Epub 2010 Apr 23. Mol Microbiol. 2010. PMID: 20444095 Review.
Cited by
- An Orphan VrgG Auxiliary Module Related to the Type VI Secretion Systems from Pseudomonas ogarae F113 Mediates Bacterial Killing.
Durán D, Vazquez-Arias D, Blanco-Romero E, Garrido-Sanz D, Redondo-Nieto M, Rivilla R, Martín M. Durán D, et al. Genes (Basel). 2023 Oct 24;14(11):1979. doi: 10.3390/genes14111979. Genes (Basel). 2023. PMID: 38002922 Free PMC article. - Comparative genomics to examine the endophytic potential of Pantoea agglomerans DAPP-PG 734.
Sulja A, Pothier JF, Blom J, Moretti C, Buonaurio R, Rezzonico F, Smits THM. Sulja A, et al. BMC Genomics. 2022 Nov 8;23(1):742. doi: 10.1186/s12864-022-08966-y. BMC Genomics. 2022. PMID: 36344949 Free PMC article. - Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome.
Durán D, Bernal P, Vazquez-Arias D, Blanco-Romero E, Garrido-Sanz D, Redondo-Nieto M, Rivilla R, Martín M. Durán D, et al. Sci Rep. 2021 Mar 11;11(1):5772. doi: 10.1038/s41598-021-85218-1. Sci Rep. 2021. PMID: 33707614 Free PMC article. - Complete Genome Sequence of Pantoea stewartii RON18713 from Brazil Nut Tree Phyllosphere Reveals Genes Involved in Plant Growth Promotion.
Rocha RT, de Almeida FM, Pappas MCR, Pappas GJ Jr, Martins K. Rocha RT, et al. Microorganisms. 2023 Jun 30;11(7):1729. doi: 10.3390/microorganisms11071729. Microorganisms. 2023. PMID: 37512901 Free PMC article. - Pantoea ananatis Genetic Diversity Analysis Reveals Limited Genomic Diversity as Well as Accessory Genes Correlated with Onion Pathogenicity.
Stice SP, Stumpf SD, Gitaitis RD, Kvitko BH, Dutta B. Stice SP, et al. Front Microbiol. 2018 Feb 13;9:184. doi: 10.3389/fmicb.2018.00184. eCollection 2018. Front Microbiol. 2018. PMID: 29491851 Free PMC article.
References
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Giford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. - DOI - PMC - PubMed
- Pukatzki S, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous