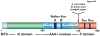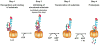Multitasking in the mitochondrion by the ATP-dependent Lon protease - PubMed (original) (raw)
Review
Multitasking in the mitochondrion by the ATP-dependent Lon protease
Sundararajan Venkatesh et al. Biochim Biophys Acta. 2012 Jan.
Abstract
The AAA(+) Lon protease is a soluble single-ringed homo-oligomer, which represents the most streamlined operational unit mediating ATP-dependent proteolysis. Despite its simplicity, the architecture of Lon proteases exhibits a species-specific diversity. Homology modeling provides insights into the structural features that distinguish bacterial and human Lon proteases as hexameric complexes from yeast Lon, which is uniquely heptameric. The best-understood functions of mitochondrial Lon are linked to maintaining proteostasis under normal metabolic conditions, and preventing proteotoxicity during environmental and cellular stress. An intriguing property of human Lon is its specific binding to G-quadruplex DNA, and its association with the mitochondrial genome in cultured cells. A fraction of Lon preferentially binds to the control region of mitochondrial DNA where transcription and replication are initiated. Here, we present an overview of the diverse functions of mitochondrial Lon, as well as speculative perspectives on its role in protein and mtDNA quality control.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
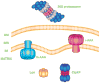
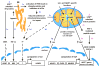
Fig. 1. ATP-dependent proteolytic machines within metazoan cells
The 26S proteasome is the only non-mitochondrial energy-dependent protease within animal cells that operates in the cytosol and nucleus. i-AAA and m-AAA are transmembrane proteases located within the mitochondrial inner membrane (IM) with their catalytic sites for ATP hydrolysis and proteolysis located within the intermembrane space (IMS) and matrix, respectively. Lon and ClpXP are soluble proteases within the matrix.
Fig. 2
(A) Domain structure and functional motifs within the primary amino acid sequence of human mitochondrial Lon. MTS - the mitochondrial targeting sequence of the Lon precursor protein binds to the mitochondrial outer membrane translocation machinery, which mediates protein import into the matrix. N domain- a highly variable amino-terminal domain proposed to function in substrate recognition and binding. AAA+ module- composed of the α/β sub-domain, which contains the Walker Box A and B motifs for ATP -binding and –hydrolysis, in addition to the small α sub-domain. P domain- the protease domain containing the serine (S) and lysine (K) residues forming the catalytic dyad at the proteolytic active sites. S. cerevisiae Lon contains a 54 amino acid charged region that is present between the AAA+ and protease modules. (B) Alignment of the primary amino acid sequences that include the Lon AAA+ module and P-domain from H. sapiens, S. cerevisiae and B. subtilis. Alignment was performed using Jotun-Hein Method. The sequence similarity within this region between H. sapiens and S. cerevisiae and B. subtilis is 58% and 49%, respectively. Walker Box motifs A and B are indicated by a blue line; the highly charged region present in yeast Lon/Pim1p is highlighted in yellow; and the residues forming the serine-lysine dyad are outlined in red.
Fig. 2
(A) Domain structure and functional motifs within the primary amino acid sequence of human mitochondrial Lon. MTS - the mitochondrial targeting sequence of the Lon precursor protein binds to the mitochondrial outer membrane translocation machinery, which mediates protein import into the matrix. N domain- a highly variable amino-terminal domain proposed to function in substrate recognition and binding. AAA+ module- composed of the α/β sub-domain, which contains the Walker Box A and B motifs for ATP -binding and –hydrolysis, in addition to the small α sub-domain. P domain- the protease domain containing the serine (S) and lysine (K) residues forming the catalytic dyad at the proteolytic active sites. S. cerevisiae Lon contains a 54 amino acid charged region that is present between the AAA+ and protease modules. (B) Alignment of the primary amino acid sequences that include the Lon AAA+ module and P-domain from H. sapiens, S. cerevisiae and B. subtilis. Alignment was performed using Jotun-Hein Method. The sequence similarity within this region between H. sapiens and S. cerevisiae and B. subtilis is 58% and 49%, respectively. Walker Box motifs A and B are indicated by a blue line; the highly charged region present in yeast Lon/Pim1p is highlighted in yellow; and the residues forming the serine-lysine dyad are outlined in red.
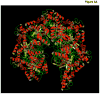
Fig. 3. Molecular modeling of human mitochondrial Lon protease
(A) The ribbon model was generated in PyMol. (B) The space filling model was generated in Schrödinger Suite. Models of the human Lon monomer containing the ATPase and protease domains were initially generated by the CPH server [121], (
http://www.cbs.dtu.dk/services/CPHmodels/
), which uses the SEGMOD tool of GeneMine (Molecular Application Group, GeneMine, Palo Alto, CA, formerly known as XLOOK) [–124]. SEGMOD divides the target sequence into short segments followed by searches for homologous structural fragments in Protein Data Bank (PDB). The fragments are then fitted onto the template structure using secondary structure alignment and multiple independent models were generated of the entire structure. The optimal structure is the average of multiple models (GeneMine Manual, pp. 425 – 426). The resulting structures were submitted to the PDB to identify protein structures with highest homology. A sequence alignment was then generated to identify conserved and defined regions of secondary structure. The secondary structural alignment demonstrated that human Lon has 43.3% identity with Bacillus subtilis Lon (Supplementary Figure 1). By contrast, human Lon showed significantly lower structural identity (36%) with Thermococcus onnurineus NA1 (Ton) Lon. Therefore, the crystal structure of B. subtilis Lon (PDB file 3M6A) [22], was employed as the template structure to model human Lon. The loops joining the secondary structures were modeled by searching for homologous structures in PDB. The side chains were then added using energy constraints. The model obtained from the CPH server was refined using Schrödinger Suite (Schrödinger Inc. LLC, New York, NY). The ‘Prepare Protein’ protocol was used to assign bond orders and valences of each atom, to add the required hydrogen atoms and to optimize H-bond and side chain orientations. The resulting monomer structure was minimized using the ‘Impact’ utility with a OPLS2005 force field. The quaternary structure model of human Lon was generated by Sybyl (Version X; Tripos Associates, St. Louis, MO). A computer program in Sybyl Programming Language (SPL) was written to incorporate a C6 symmetry related operation with energy constraints that limited the steric clash to a distance of 0.5 Å between two side chain atoms at the interface of adjacent monomers in the complex. In the event of steric clashes, energy minimization of the side chains was performed using AMBER force field and Gesteiger-Marshelili charges [125, 126]. The final model was further subject to ‘Protein Preparation’ Wizard’ tool of Schrödinger Suite. A similar protocol was used to generate the model for yeast mitochondrial Lon protease.
Fig. 3. Molecular modeling of human mitochondrial Lon protease
(A) The ribbon model was generated in PyMol. (B) The space filling model was generated in Schrödinger Suite. Models of the human Lon monomer containing the ATPase and protease domains were initially generated by the CPH server [121], (
http://www.cbs.dtu.dk/services/CPHmodels/
), which uses the SEGMOD tool of GeneMine (Molecular Application Group, GeneMine, Palo Alto, CA, formerly known as XLOOK) [–124]. SEGMOD divides the target sequence into short segments followed by searches for homologous structural fragments in Protein Data Bank (PDB). The fragments are then fitted onto the template structure using secondary structure alignment and multiple independent models were generated of the entire structure. The optimal structure is the average of multiple models (GeneMine Manual, pp. 425 – 426). The resulting structures were submitted to the PDB to identify protein structures with highest homology. A sequence alignment was then generated to identify conserved and defined regions of secondary structure. The secondary structural alignment demonstrated that human Lon has 43.3% identity with Bacillus subtilis Lon (Supplementary Figure 1). By contrast, human Lon showed significantly lower structural identity (36%) with Thermococcus onnurineus NA1 (Ton) Lon. Therefore, the crystal structure of B. subtilis Lon (PDB file 3M6A) [22], was employed as the template structure to model human Lon. The loops joining the secondary structures were modeled by searching for homologous structures in PDB. The side chains were then added using energy constraints. The model obtained from the CPH server was refined using Schrödinger Suite (Schrödinger Inc. LLC, New York, NY). The ‘Prepare Protein’ protocol was used to assign bond orders and valences of each atom, to add the required hydrogen atoms and to optimize H-bond and side chain orientations. The resulting monomer structure was minimized using the ‘Impact’ utility with a OPLS2005 force field. The quaternary structure model of human Lon was generated by Sybyl (Version X; Tripos Associates, St. Louis, MO). A computer program in Sybyl Programming Language (SPL) was written to incorporate a C6 symmetry related operation with energy constraints that limited the steric clash to a distance of 0.5 Å between two side chain atoms at the interface of adjacent monomers in the complex. In the event of steric clashes, energy minimization of the side chains was performed using AMBER force field and Gesteiger-Marshelili charges [125, 126]. The final model was further subject to ‘Protein Preparation’ Wizard’ tool of Schrödinger Suite. A similar protocol was used to generate the model for yeast mitochondrial Lon protease.
Fig. 4. General paradigm for the recognition and degradation of protein substrates by the Lon protease
Step 1 - recognition and binding of a structural feature or sequence within a protein substrate. **Step 2**- unfolding of structured protein substrates by the AAA+ domains of the holoenzyme, which requires ATP –binding and -hydrolysis. Unfolded substrates or peptides bypass this step. **Step 3**- translocation of unfolded polypeptides or short peptide sequences into the degradation chamber, which also requires ATP –binding and -hydrolysis. Step 4- peptide bond cleavage resulting in the generation of small peptide products. Reiterative rounds of substrate unfolding and translocation are required to degrade substrates completely.
Fig. 5. Regulation of Lon and ClpXP in the ER and mitochondrial unfolded protein responses
(1) Consequences of the unfolded protein response in the ER (UPRER)(1a) The accumulation of unfolded or misfolded proteins in the ER lumen activates PERK (protein kinase RNA-like endoplasmic reticulum kinase), which phosphorylates and thereby inhibits the translation initiation factor eIF2α. (1b) A block in eIF2α-dependent translation leads to the selective reduction of cytosolic protein synthesis, which includes proteins imported into mitochondria. (1c) The reduction in proteins imported from the cytosol may result in a stoichiometric imbalance of mitochondrial proteins that must assemble to form functional complexes. (1d) Signals originating from the cytosol or the mitochondrion may transcriptionally up-regulate the LONP1 gene by an unknown mechanism. (1e) Alternatively, accumulation of unfolded proteins in the ER directly activates the translation of the transcription factor XBP1 (X-box binding protein 1), which translocates to the nucleus and may activate LONP1 expression by binding to a putative UPRE (unfolded protein response element), which has yet to be identified. (1f) Hypoxia is a physiological inducer of ER stress that has been shown to up-regulate Lon expression by the activation of HIF-1α (hypoxia-inducible factor 1α), which binds to HREs (hypoxia response element) within the promoter of LONP1. (1g) The resulting increase of Lon may function to re-establish mitochondrial homeostasis. (2) Consequences of the unfolded protein response in the mitochondria (UPRMT). (2a) Accumulation of unfolded or misfolded proteins that tax the protein quality control capacity of mitochondria lead to the UPRMT, which is initiated by ClpXP-dependent degradation of protein substrates thereby generating peptides that are effluxed from the matrix to the cytosol by the HAF-1 transporter located in the mitochondrial inner membrane. (2b) The effluxed peptides activate the transcription factors (ZC376.7 and possibly the CHOP/C/EBPβ heterodimer), which then translocate into the nucleus, binding to MURE and CHOP sequences up-regulating the expression of UPRMT related genes encoding ClpP, mtHsp70, mtHsp60, mtHsp10, and mtDnaJ. (2c) The increased expression of ClpP relieves mitochondrial stress and re-establishes mitochondrial homeostasis.
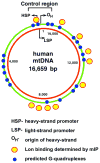
Fig. 6. Relative locations within the mitochondrial genome of predicted G-quadruplexes and Lon binding
The positions of G-quadruplexes within the heavy-strand of human mtDNA (NC_001807) were predicted using Quadfinder Version 1 (
http://miracle.igib.res.in/quadfinder/
) [16]. The regions of mtDNA bound by Lon in cultured HeLa cells was determined by mtDNA immunoprecipitation (mIP) [15].
Similar articles
- Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease.
Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Lu B, et al. Mol Cell. 2013 Jan 10;49(1):121-32. doi: 10.1016/j.molcel.2012.10.023. Epub 2012 Nov 29. Mol Cell. 2013. PMID: 23201127 Free PMC article. - The role of Lon-mediated proteolysis in the dynamics of mitochondrial nucleic acid-protein complexes.
Kunová N, Ondrovičová G, Bauer JA, Bellová J, Ambro Ľ, Martináková L, Kotrasová V, Kutejová E, Pevala V. Kunová N, et al. Sci Rep. 2017 Apr 4;7(1):631. doi: 10.1038/s41598-017-00632-8. Sci Rep. 2017. PMID: 28377575 Free PMC article. - Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
Yang J, Song AS, Wiseman RL, Lander GC. Yang J, et al. J Biol Chem. 2022 Mar;298(3):101694. doi: 10.1016/j.jbc.2022.101694. Epub 2022 Feb 7. J Biol Chem. 2022. PMID: 35143841 Free PMC article. - Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates.
Lee I, Suzuki CK. Lee I, et al. Biochim Biophys Acta. 2008 May;1784(5):727-35. doi: 10.1016/j.bbapap.2008.02.010. Epub 2008 Mar 5. Biochim Biophys Acta. 2008. PMID: 18359303 Free PMC article. Review. - Towards the control of intracellular protein turnover: mitochondrial Lon protease inhibitors versus proteasome inhibitors.
Bayot A, Basse N, Lee I, Gareil M, Pirotte B, Bulteau AL, Friguet B, Reboud-Ravaux M. Bayot A, et al. Biochimie. 2008 Feb;90(2):260-9. doi: 10.1016/j.biochi.2007.10.010. Epub 2007 Oct 25. Biochimie. 2008. PMID: 18021745 Review.
Cited by
- Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Shin M, Watson ER, Song AS, Mindrebo JT, Novick SJ, Griffin PR, Wiseman RL, Lander GC. Shin M, et al. Nat Commun. 2021 May 28;12(1):3239. doi: 10.1038/s41467-021-23495-0. Nat Commun. 2021. PMID: 34050165 Free PMC article. - Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives.
Nordgren M, Wang B, Apanasets O, Fransen M. Nordgren M, et al. Front Physiol. 2013 Jun 14;4:145. doi: 10.3389/fphys.2013.00145. eCollection 2013. Front Physiol. 2013. PMID: 23785334 Free PMC article. - The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives.
Bernstein SH, Venkatesh S, Li M, Lee J, Lu B, Hilchey SP, Morse KM, Metcalfe HM, Skalska J, Andreeff M, Brookes PS, Suzuki CK. Bernstein SH, et al. Blood. 2012 Apr 5;119(14):3321-9. doi: 10.1182/blood-2011-02-340075. Epub 2012 Feb 8. Blood. 2012. PMID: 22323447 Free PMC article. - Mitochondrial cereblon functions as a Lon-type protease.
Kataoka K, Nakamura C, Asahi T, Sawamura N. Kataoka K, et al. Sci Rep. 2016 Jul 15;6:29986. doi: 10.1038/srep29986. Sci Rep. 2016. PMID: 27417535 Free PMC article. - Activated PyK2 and Its Associated Molecules Transduce Cellular Signaling from the Cancerous Milieu for Cancer Metastasis.
Lee D, Hong JH. Lee D, et al. Int J Mol Sci. 2022 Dec 7;23(24):15475. doi: 10.3390/ijms232415475. Int J Mol Sci. 2022. PMID: 36555115 Free PMC article. Review.
References
- Bota DA, Davies KJ. Protein degradation in mitochondria: implications for oxidative stress, aging and disease: a novel etiological classification of mitochondrial proteolytic disorders. Mitochondrion. 2001;1:33–49. - PubMed
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. - PubMed
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. - PubMed
- Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 NS067668-01A1/NS/NINDS NIH HHS/United States
- R01 GM084039-02/GM/NIGMS NIH HHS/United States
- R21NS067668/NS/NINDS NIH HHS/United States
- R01 GM061095/GM/NIGMS NIH HHS/United States
- R01GM084039/GM/NIGMS NIH HHS/United States
- R21 NS067668/NS/NINDS NIH HHS/United States
- R01 GM061095-05/GM/NIGMS NIH HHS/United States
- R01 GM084039/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases