Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition - PubMed (original) (raw)
Comparative Study
. 2011 Dec 27;108(52):E1475-83.
doi: 10.1073/pnas.1117988108. Epub 2011 Nov 28.
Affiliations
- PMID: 22123957
- PMCID: PMC3248478
- DOI: 10.1073/pnas.1117988108
Comparative Study
Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition
Jason R Rock et al. Proc Natl Acad Sci U S A. 2011.
Abstract
There are currently few treatment options for pulmonary fibrosis. Innovations may come from a better understanding of the cellular origin of the characteristic fibrotic lesions. We have analyzed normal and fibrotic mouse and human lungs by confocal microscopy to define stromal cell populations with respect to several commonly used markers. In both species, we observed unexpected heterogeneity of stromal cells. These include numerous cells with molecular and morphological characteristics of pericytes, implicated as a source of myofibroblasts in other fibrotic tissues. We used mouse genetic tools to follow the fates of specific cell types in the bleomcyin-induced model of pulmonary fibrosis. Using inducible transgenic alleles to lineage trace pericyte-like cells in the alveolar interstitium, we show that this population proliferates in fibrotic regions. However, neither these cells nor their descendants express high levels of the myofibroblast marker alpha smooth muscle actin (Acta2, aSMA). We then used a Surfactant protein C-CreER(T2) knock-in allele to follow the fate of Type II alveolar cells (AEC2) in vivo. We find no evidence at the cellular or molecular level for epithelial to mesenchymal transition of labeled cells into myofibroblasts. Rather, bleomycin accelerates the previously reported conversion of AEC2 into AEC1 cells. Similarly, epithelial cells labeled with our Scgb1a1-CreER allele do not give rise to fibroblasts but generate both AEC2 and AEC1 cells in response to bleomycin-induced lung injury. Taken together, our results show a previously unappreciated heterogeneity of cell types proliferating in fibrotic lesions and exclude pericytes and two epithelial cell populations as the origin of myofibroblasts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
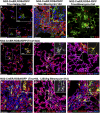
Heterogeneity of stromal cells in normal and fibrotic mouse lungs. Sections of normal mouse lungs (A, B, D, E, G, H, J, and K) and fibrotic lungs collected 21 d after the intratracheal administration of 1.25 U/kg bleomycin (C, F, I, and L) were stained with antibodies against stromal cell markers. Single images from representative confocal stacks are shown. In some cases, one marker (noted in parentheses) is omitted from high-magnification Insets for clarity. (A–C) aSMA (red), S100a4 (green), and vimentin (purple). Arrows in B, G, and I mark vimentin-positive macrophage identified by shape and location within the alveolar space. (C) Note increase in S100a4-positive cells in fibrotic regions. Some of these cells coexpress vimentin (Inset Upper, white) and CD45, a marker of hematopoietic cells (Inset Lower, yellow). (Note that this image is from a different fibrotic lung.) (D –F) Pdgfrb (red), desmin (green), and aSMA (purple). Note coexpression of desmin and aSMA (white) in smooth muscle cells around blood vessels and bronchioles. Desmin is also expressed in Pdgfrb-positive pericytes (D –F Insets, yellow) and Pdgfrb-negative fibroblasts (D and E Insets, green) in the alveoli. Desmin-positive cells, with and without coexpression of Pdgfrb, are abundant within fibrotic foci. (G–I) aSMA (red), NG2 (Cspg4; green), and vimentin (purple). NG2-positive cells coexpress aSMA (yellow) around blood vessels, but the abundant NG2-positive pericyte-like cells within the alveolar interstitium do not express aSMA. Fibrotic areas contain large numbers of NG2-positive cells, most of which do not express aSMA (yellow). (J–L) NG2 (green) and PECAM (CD31; purple). Pericyte-like cells are in close association with pulmonary microvasculature, even in areas of fibrosis. Dashed boxed region of each image is shown at higher magnification in Insets. J Inset shows NG2 (green) and PECAM (red) at high magnification for clarity. br, bronchiole; bv, blood vessel. (Scale bars: A, D, G, and J, 50 μm; Insets, 20 μm.)
Fig. 2.
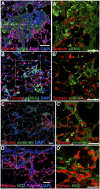
NG2-CreER labels pericyte-like interstitial cells that proliferate in pulmonary fibrosis. (A and B) NG2-CreER;ROSA-fGFP double heterozygous mice were given four doses of tamoxifen followed 4 d later with intratracheal (A) saline or (B) 1.25 U/kg bleomycin; 14 d later, sections were stained with antibodies against GFP and aSMA (red). Note that the lineage label does not colocalize with aSMA in normal or fibrotic lungs. (C) NG2-CreER;ROSA-fGFP double heterozygous mice were given tamoxifen and bleomycin as above. BrdU was given intraperitoneally 3 h before animals were killed 14 d later. Sections were stained with antibodies against GFP (green, lineage label) and BrdU (red, proliferative cells). The percentage of proliferating lineage-labeled pericyte-like cells (arrows) increases from 0.51% in controls (n = 3 mice) to 2.46% in fibrotic regions after bleomycin (n = 4 mice). (D –I) NG2-CreER;ROSA-fGFP double heterozygous mice were given four doses of tamoxifen followed 4 d later with intratracheal (D –F) saline or (G–I) 1.25 U/kg bleomycin; 21 d later, sections were stained with antibodies against GFP (green, lineage label), (D and G) NG2 (red) and aSMA (purple), (E and H) PECAM (red) and NG2 (purple), or (F and I) Pdgfrb (red) and Pdpn (purple). (D –F) Lineage-labeled pericyte-like cells (D, yellow) are intimately associated with the pulmonary microvasculature and express (E) NG2 and (F) Pdgfrb. (G–I) In fibrotic regions following bleomycin, the lineage-labeled pericyte-like population (yellow) expands and continues to express (G and H) NG2 and (I) Pdgfrb. The dashed boxed region of each image is shown at higher magnification in Insets. In some cases, one marker (noted in parentheses) is omitted from high-magnification Insets for clarity. br, bronchiole; bv, blood vessel. (Scale bars: A, B, C, D, and G, 50 μm; Insets A, D, and G, 20 μm.)
Fig. 3.
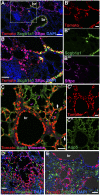
AEC2 cells give rise to AEC1 under steady-state conditions and in response to bleomycin. (A) Adult Sftpc-CreERT2;ROSA-Tomato mice were given four doses of tamoxifen to induce the heritable expression of the RFP variant tdTomato in AEC2 cells. This regimen labels ∼84.2 ± 4% of Sftpc+ AEC2 when sections are analyzed 4 d after the final dose of tamoxifen (n = 3 mice). (B) The proportion of lineage-labeled cells scored as AEC1 based on morphology and expression of Aqp5 or Pdpn was determined 5, 10, and 21 d after the intratracheal administration of 1.25 U/kg bleomycin or saline control. Data shown are means ± SD and n = 3 mice for each group. (C and D) Sixty days after bleomycin, the proportion of lineage-labeled Aqp5- and Pdpn-positive AEC1 (arrows) was increased compared with saline-treated controls (E) or relatively unaffected regions of bleomycin-treated lungs (C, right side). In C′, C″, and D′, DAPI and red and/or green channels are omitted for clarity. (Scale bars: A–E, 50 μm; D′, 20 μm.)
Fig. 4.
AEC2 cells do not directly contribute to pulmonary fibrosis by EMT. Sftpc-CreERT2;ROSA-Tomato double heterozygous mice were given four doses of tamoxifen followed 4–10 d later with 1.25 U/kg intratracheal bleomycin. After 21 d, we imaged endogenous Tomato lineage label and immunofluorescence against (A) aSMA (green) and Aqp5 (purple), (B) S100a4 (green), (C) vimentin (green), and (D) NG2 (green) and Pdgfrb (purple). Note that some Aqp5 AEC1 cells are lineage-labeled (A) but that there is no colocalization of lineage label with stromal markers. Red and green channels from boxed regions from A–D are shown at higher magnification in A′–D′. (Scale bars: A, A′, B, B′, C′, and D, 50 μm; C, 100 μm; D′, 25 μm.)
Fig. 5.
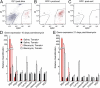
Gene expression in freshly sorted cells from normal and fibrotic lungs. Adult Sftpc-CreERT2;ROSA-Tomato mice were given four doses of tamoxifen followed 4–10 d later by intratracheal saline or 1.25 U/kg bleomycin. (A) After 10 or 21 d, lungs were dissociated and sorted into Tomato-positive (P3; lineage labeled) and -negative (P4; unlabeled) populations by FACS. Resorting of (B) positive and (C) negative populations shows purity. Expression of Sftpc (AEC2), Pdpn (AEC1), aSMA, Col1a1, versican, vimentin, S100a4, and NG2 relative to GAPDH and Tbp was measured by qPCR in Tomato-positive (red bars) and -negative (black bars) populations (D) 10 or (E) 21 d after treatment with saline (solid bars) or bleomycin (open bars). The expression level of each gene in Tomato-negative cells from saline-treated lungs was set equal to one for comparison. Combined data from three independent experiments (each data point performed in triplicate) are shown, and error bars indicate 95% confidence interval. Note that Sftpc expression is highest in the Tomato-positive populations. The level of Pdpn is increased in the Tomato-positive population 21 d after bleomycin, consistent with the derivation of AEC1 from lineage-labeled AEC2 progenitors. Sftpc is modestly increased in the Tomato-negative population after bleomycin, presumably because AEC2 can derive from an Sftpc-negative unlabeled progenitor under these conditions. The expression of aSMA, Col1a1, versican, vimentin, S100a4, and NG2 is restricted to the Tomato-negative, predominantly stromal populations in normal and fibrotic lungs.
Fig. 6.
Scgb1a1-positive epithelial cells give rise to AEC2 and AEC1 cells, but not stromal cells, in pulmonary fibrosis. Scgb1a1-CreER;ROSA-Tomato double heterozygous mice were given one or four doses of tamoxifen followed 4–10 d later with 2.5 U/kg intratracheal bleomycin. After 21 d, sections were stained with antibodies against RFP (red, Tomato lineage label) and (A and B) Scgb1a1 (green) and Sftpc (purple), (C) Aqp5 (green) and vimentin (purple), (D) vimentin (green), or (E) S100A4 (green). (A and B) Note that bronchiolar Scgb1a1-positive cells and alveolar Scgb1a1-negative cells are lineage labeled (red), suggesting that they are derived from an Scgb1a1-positive progenitor. Many of the lineage-labeled alveolar cells express (A and B) the AEC2 marker Sftpc or (C) AEC1 marker Aqp5 (yellow, arrows). In contrast, there is no colocalization of the lineage label with the stromal markers (C and D) vimentin or (E) S100a4. Boxed region in A shown at higher magnification in B. Single channels from B and C are shown independently in B′–B″′ and C′–C″′ for clarity. br, bronchiole. (Scale bars: A, 100 μm; B, B′, C, C′, D, and E, 50 μm.)
Fig. P1.
Schematic depiction of alveoli in a normal mouse lung (Left) and remodeling in a region of bleomycin-induced fibrosis (Right). In the normal lung, lineage-labeled AEC2 gives rise to AEC1 (black arrow), supporting the classical view of AEC2 cells as alveolar epithelial progenitor. The rate of this conversion is enhanced in response to bleomycin injury, but lineage-labeled epithelial cells do not give rise to fibroblasts through epithelial to mesenchymal transition. After bleomycin-induced lung injury, Scgb1a1+ lineage-labeled cells also proliferate and generate AEC2 and AEC1 cells. In fibrotic lungs, lineage-labeled pericytes proliferate but are not a major source of aSMA-positive myofibroblasts.
Similar articles
- Detection of epithelial to mesenchymal transition in airways of a bleomycin induced pulmonary fibrosis model derived from an alpha-smooth muscle actin-Cre transgenic mouse.
Wu Z, Yang L, Cai L, Zhang M, Cheng X, Yang X, Xu J. Wu Z, et al. Respir Res. 2007 Jan 7;8(1):1. doi: 10.1186/1465-9921-8-1. Respir Res. 2007. PMID: 17207287 Free PMC article. - The increase of microRNA-21 during lung fibrosis and its contribution to epithelial-mesenchymal transition in pulmonary epithelial cells.
Yamada M, Kubo H, Ota C, Takahashi T, Tando Y, Suzuki T, Fujino N, Makiguchi T, Takagi K, Suzuki T, Ichinose M. Yamada M, et al. Respir Res. 2013 Sep 24;14(1):95. doi: 10.1186/1465-9921-14-95. Respir Res. 2013. PMID: 24063588 Free PMC article. - CXCL4 drives fibrosis by promoting several key cellular and molecular processes.
Affandi AJ, Carvalheiro T, Ottria A, de Haan JJ, Brans MAD, Brandt MM, Tieland RG, Lopes AP, Fernández BM, Bekker CPJ, van der Linden M, Zimmermann M, Giovannone B, Wichers CGK, Garcia S, de Kok M, Stifano G, Xu YJ, Kowalska MA, Waasdorp M, Cheng C, Gibbs S, de Jager SCA, van Roon JAG, Radstake TRDJ, Marut W. Affandi AJ, et al. Cell Rep. 2022 Jan 4;38(1):110189. doi: 10.1016/j.celrep.2021.110189. Cell Rep. 2022. PMID: 34986347 - Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders.
Piera-Velazquez S, Li Z, Jimenez SA. Piera-Velazquez S, et al. Am J Pathol. 2011 Sep;179(3):1074-80. doi: 10.1016/j.ajpath.2011.06.001. Epub 2011 Jul 19. Am J Pathol. 2011. PMID: 21763673 Free PMC article. Review. - Genesis of the myofibroblast in lung injury and fibrosis.
Phan SH. Phan SH. Proc Am Thorac Soc. 2012 Jul;9(3):148-52. doi: 10.1513/pats.201201-011AW. Proc Am Thorac Soc. 2012. PMID: 22802289 Free PMC article. Review.
Cited by
- Inhibition of Aurora Kinase B attenuates fibroblast activation and pulmonary fibrosis.
Kasam RK, Ghandikota S, Soundararajan D, Reddy GB, Huang SK, Jegga AG, Madala SK. Kasam RK, et al. EMBO Mol Med. 2020 Sep 7;12(9):e12131. doi: 10.15252/emmm.202012131. Epub 2020 Aug 6. EMBO Mol Med. 2020. PMID: 32761869 Free PMC article. - Conceptual approaches to lung injury and repair.
Zemans RL, Henson PM, Henson JE, Janssen WJ. Zemans RL, et al. Ann Am Thorac Soc. 2015 Mar;12 Suppl 1(Suppl 1):S9-15. doi: 10.1513/AnnalsATS.201408-402MG. Ann Am Thorac Soc. 2015. PMID: 25830855 Free PMC article. Review. - Cellular mechanisms of tissue fibrosis. 3. Novel mechanisms of kidney fibrosis.
Campanholle G, Ligresti G, Gharib SA, Duffield JS. Campanholle G, et al. Am J Physiol Cell Physiol. 2013 Apr 1;304(7):C591-603. doi: 10.1152/ajpcell.00414.2012. Epub 2013 Jan 16. Am J Physiol Cell Physiol. 2013. PMID: 23325411 Free PMC article. Review. - Transforming growth factor-β in tissue fibrosis.
Frangogiannis N. Frangogiannis N. J Exp Med. 2020 Feb 13;217(3):e20190103. doi: 10.1084/jem.20190103. Print 2020 Mar 2. J Exp Med. 2020. PMID: 32997468 Free PMC article. Review. - Regulation of idiopathic pulmonary fibrosis: a cross-talk between TGF-β signaling and MicroRNAs.
Wang S, Yu H, Liu S, Liu Y, Gu X. Wang S, et al. Front Med (Lausanne). 2024 Sep 25;11:1415278. doi: 10.3389/fmed.2024.1415278. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39386739 Free PMC article. Review.
References
- King TE, Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011 10.1016/S0140-6736(11)60052-4. - PubMed
- Plantier L, et al. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66:651–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32HL007538/HL/NHLBI NIH HHS/United States
- F32 HL102920/HL/NHLBI NIH HHS/United States
- R01 HL071303/HL/NHLBI NIH HHS/United States
- HL071303/HL/NHLBI NIH HHS/United States
- R37 HL071303/HL/NHLBI NIH HHS/United States
- R01 HL060539/HL/NHLBI NIH HHS/United States
- HL102920/HL/NHLBI NIH HHS/United States
- T32 HL007538/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous