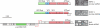Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere - PubMed (original) (raw)
Review
Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere
Mart Krupovic et al. Microbiol Mol Biol Rev. 2011 Dec.
Abstract
Prokaryotes, bacteria and archaea, are the most abundant cellular organisms among those sharing the planet Earth with human beings (among others). However, numerous ecological studies have revealed that it is actually prokaryotic viruses that predominate on our planet and outnumber their hosts by at least an order of magnitude. An understanding of how this viral domain is organized and what are the mechanisms governing its evolution is therefore of great interest and importance. The vast majority of characterized prokaryotic viruses belong to the order Caudovirales, double-stranded DNA (dsDNA) bacteriophages with tails. Consequently, these viruses have been studied (and reviewed) extensively from both genomic and functional perspectives. However, albeit numerous, tailed phages represent only a minor fraction of the prokaryotic virus diversity. Therefore, the knowledge which has been generated for this viral system does not offer a comprehensive view of the prokaryotic virosphere. In this review, we discuss all families of bacterial and archaeal viruses that contain more than one characterized member and for which evolutionary conclusions can be attempted by use of comparative genomic analysis. We focus on the molecular mechanisms of their genome evolution as well as on the relationships between different viral groups and plasmids. It becomes clear that evolutionary mechanisms shaping the genomes of prokaryotic viruses vary between different families and depend on the type of the nucleic acid, characteristics of the virion structure, as well as the mode of the life cycle. We also point out that horizontal gene transfer is not equally prevalent in different virus families and is not uniformly unrestricted for diverse viral functions.
Figures
Fig. 1.
Viruses of the order Caudovirales. Transmission electron micrographs of a T4-like virus, HK97, and P22 are shown to represent the families Myoviridae, Siphoviridae, and Podoviridae, respectively. The three-dimensional structures of the major capsid proteins of the corresponding viruses are shown underneath the electron micrographs. Protein Data Bank (PDB) accession numbers are 1YUE for T4 gp24 (82), 1OHG for HK97 gp5 (288), and 2XYY for P22 gp5 (43). (The electron micrograph of a T4-like virus is courtesy of Damien Maura and Laurent Debarbieux, Institut Pasteur. The micrographs of HK97 and P22 are reproduced from reference .) The bar is the same for all three micrographs, 50 nm.
Fig. 2.
Mosaic organization of the tailed dsDNA virus genomes. Homologous genes in cassettes are colored similarly; analogous but not homologous genes are shown in orange. To a first approximation, the head genes stay together as a single mosaic module, as do the tail genes. It should be noted, however, that the two modules are not strictly linked and can be exchanged between distantly related viruses. The early genes (right end of the map for all but phage Mu) are mosaic on a finer scale, with individual genes typically representing mosaic modules. (Reproduced from reference .)
Fig. 3.
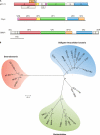
Genomic organization and phylogenetic relationship between members of the family Microviridae. (A) Genome maps of representative members of three distinct groups of viruses within the Microviridae: Escherichia coli-infecting φX174 (genus Microvirus), Chlamydia psittaci-infecting Chp1 (genus Chlamydiamicrovirus, subfamily Gokushovirinae), and provirus BMV1 integrated into the genome of Bacteroides sp. strain 2-2-4. Homologous genes are colored similarly. attL and attR, left and right attachment sites, respectively. (B) Unrooted maximum likelihood phylogenetic tree of the φX174 gpF-like major capsid proteins representing the relationship between enterobacterium-infecting microviruses, obligate intracellular bacterium-infecting gokushoviruses, and putative proviruses integrated into the genomes of different species of the phylum Bacteroidetes. Putative marine microviruses SARssφ1 and SARssφ2 (273) cluster together with gokushoviruses. (For accession numbers of the proteins that were used to generate the phylogeny, please refer to reference .)
Fig. 4.
Mosaic organization of ssDNA filamentous virus genomes. (A) Negative-contrast transmission electron micrograph of a Pseudomonas aeruginosa-infecting filamentous virus, phi05_2302 (88). (The micrograph is courtesy of Aušra Domanska.) Bar, 200 nm. (B) Homologous genes in cassettes are colored similarly; orthologous gene cassettes with various degrees of similarity are represented by various shades of the same color, whereas nonorthologous replacements are denoted by differently colored cassettes. The darker regions in genes I and IV of inoviruses IKe and I2-2 indicate homologous recombination events with M13/fd-like sequences. (Panel B is reproduced from reference .)
Fig. 5.
Genomic organizations found in ssRNA viruses of the family Leviviridae. Note the various locations of the lysis protein-coding gene. During Qβ infection, cell lysis is performed by the maturation protein (19).
Fig. 6.
Negative-contrast transmission electron micrographs of virions from different families of crenarchaeal viruses, including the Fuselloviridae (left, SSV1; right, SSV6), Rudiviridae (SIRV1), Lipothrixviridae (AFV1), Globuloviridae (PSV), and the unclassified virus STIV2. Bars, 100 nm.
Fig. 7.
Genomic relationship between linear archaeal viruses of the families Rudiviridae and Lipothrixviridae. Genes shared by Sulfolobus islandicus rod-shaped virus 1 (SIRV1) (Rudiviridae) and the lipothrixviruses Sulfolobus islandicus filamentous virus (SIFV) and Acidianus filamentous virus 1 (AFV1) are shaded blue. Genes restricted to the virus pairs SIRV1-SIFV, SIRV1-AFV1, and SIFV-AFV1 are shown in red, yellow, and green, respectively.
Fig. 8.
Genomic relationship between euryarchaeal pleomorphic and spindle-shaped viruses with different nucleic acid types. The electron micrographs and the types of nucleic acids comprising the genomes of the corresponding viruses are indicated on the right. Boxes above and below the vertical lines represent genes transcribed from left to right and from right to left, respectively (with kinked arrows indicating the direction of transcription). Genes shared by the three viruses Halorubrum pleomorphic virus 1 (HRPV-1), Haloarcula hispanica pleomorphic virus 1 (HHPV-1), and haloarchaeal virus His2 are shown in red, while those restricted to HRPV-1 and HHPV-1 are in blue. Genes encoding genome replication proteins are in green. RCR Rep, rolling-circle replication initiation proteins; p-p DNA Pol B, protein-primed family B DNA polymerase. Note that RCR initiation proteins of HRPV-1 and HHPV-1 share only 9% pairwise identity (236). (The HRPV-1 micrograph is reproduced from reference with permission from John Wiley & Sons. The HHPV-1 micrograph is reproduced from reference . The His2 micrograph is reproduced from reference with permission from Elsevier.) Bars, 100 nm.
Fig. 9.
Comparison of haloarchaeal virus SH1 and bacterial virus P23-77. Surface representations of the two virions are viewed along an icosahedral 3-fold symmetry axis. Symmetry axes are indicated with a white ellipse (2-fold), a triangle (3-fold), and a pentagon (5-fold). Symmetry-related capsomers are shown in the same color. The triangulation number (T) describes the geometrical arrangement of the capsomers and is the same for the SH1 and P23-77 capsids (T = 28). (The capsid representations are reproduced from reference with permission from Elsevier.) Genomic regions encoding homologous ATPases and the two major capsid proteins (MCP1 and MCP2) of SH1 and P23-77 are depicted above their respective capsid representations.
Fig. 10.
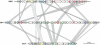
Genomic relationship between different groups of mobile genetic elements. Genomic modularity is characteristic of different types of mobile genetic elements, such as viruses, plasmids, and integrating or transposing elements. Transitions from one type of an element to another (e.g., from a plasmid to a virus) appear to have occurred on multiple independent occasions during evolution by the acquisition or loss of genes that are specific to a given group of mobile elements. The genes that are dispensable for a given type of mobile elements are shown in parentheses. CP, Rep, and Int, genes for capsid, replication, and recombination (either integrases or transposases) proteins, respectively; att, attachment sites, which mark the borders of an integrated element.
Fig. 11.
Distribution of tailed bacterial viruses according to the taxonomic grouping of their hosts. Numerical values next to the names of bacterial phyla represent the exact numbers of genome sequences available for viruses infecting bacteria from a corresponding phylum. The numbers do not include the genome sequences available for closely related viruses that are considered different isolates of the same virus strain. The data were obtained from GenBank (June 2011).
Similar articles
- Bipartite Network Analysis of the Archaeal Virosphere: Evolutionary Connections between Viruses and Capsidless Mobile Elements.
Iranzo J, Koonin EV, Prangishvili D, Krupovic M. Iranzo J, et al. J Virol. 2016 Nov 28;90(24):11043-11055. doi: 10.1128/JVI.01622-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681128 Free PMC article. - Viruses of archaea: Structural, functional, environmental and evolutionary genomics.
Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. Krupovic M, et al. Virus Res. 2018 Jan 15;244:181-193. doi: 10.1016/j.virusres.2017.11.025. Epub 2017 Nov 22. Virus Res. 2018. PMID: 29175107 Free PMC article. Review. - Orthologous gene clusters and taxon signature genes for viruses of prokaryotes.
Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. Kristensen DM, et al. J Bacteriol. 2013 Mar;195(5):941-50. doi: 10.1128/JB.01801-12. Epub 2012 Dec 7. J Bacteriol. 2013. PMID: 23222723 Free PMC article. - Vast diversity of prokaryotic virus genomes encoding double jelly-roll major capsid proteins uncovered by genomic and metagenomic sequence analysis.
Yutin N, Bäckström D, Ettema TJG, Krupovic M, Koonin EV. Yutin N, et al. Virol J. 2018 Apr 10;15(1):67. doi: 10.1186/s12985-018-0974-y. Virol J. 2018. PMID: 29636073 Free PMC article. - Exploring the prokaryotic virosphere.
Comeau AM, Hatfull GF, Krisch HM, Lindell D, Mann NH, Prangishvili D. Comeau AM, et al. Res Microbiol. 2008 Jun;159(5):306-13. doi: 10.1016/j.resmic.2008.05.001. Epub 2008 May 29. Res Microbiol. 2008. PMID: 18639443 Review.
Cited by
- Metatranscriptomic Identification of Diverse and Divergent RNA Viruses in Green and Chlorarachniophyte Algae Cultures.
Charon J, Marcelino VR, Wetherbee R, Verbruggen H, Holmes EC. Charon J, et al. Viruses. 2020 Oct 19;12(10):1180. doi: 10.3390/v12101180. Viruses. 2020. PMID: 33086653 Free PMC article. - Insights into head-tailed viruses infecting extremely halophilic archaea.
Pietilä MK, Laurinmäki P, Russell DA, Ko CC, Jacobs-Sera D, Butcher SJ, Bamford DH, Hendrix RW. Pietilä MK, et al. J Virol. 2013 Mar;87(6):3248-60. doi: 10.1128/JVI.03397-12. Epub 2013 Jan 2. J Virol. 2013. PMID: 23283946 Free PMC article. - Virus world as an evolutionary network of viruses and capsidless selfish elements.
Koonin EV, Dolja VV. Koonin EV, et al. Microbiol Mol Biol Rev. 2014 Jun;78(2):278-303. doi: 10.1128/MMBR.00049-13. Microbiol Mol Biol Rev. 2014. PMID: 24847023 Free PMC article. Review. - Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses.
Krupovic M, Koonin EV. Krupovic M, et al. Sci Rep. 2014 Jun 18;4:5347. doi: 10.1038/srep05347. Sci Rep. 2014. PMID: 24939392 Free PMC article. - A virus of hyperthermophilic archaea with a unique architecture among DNA viruses.
Rensen EI, Mochizuki T, Quemin E, Schouten S, Krupovic M, Prangishvili D. Rensen EI, et al. Proc Natl Acad Sci U S A. 2016 Mar 1;113(9):2478-83. doi: 10.1073/pnas.1518929113. Epub 2016 Feb 16. Proc Natl Acad Sci U S A. 2016. PMID: 26884161 Free PMC article.
References
- Abrescia N. G., et al. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68–74 - PubMed
- Abrescia N. G., et al. 2008. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell 31:749–761 - PubMed
- Ackermann H. W. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 - PubMed
- Agirrezabala X., et al. 2007. Quasi-atomic model of bacteriophage T7 procapsid shell: insights into the structure and evolution of a basic fold. Structure 15:461–472 - PubMed
- Ahn D. G., et al. 2006. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology 351:280–290 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources