Antibiotics in feed induce prophages in swine fecal microbiomes - PubMed (original) (raw)
Antibiotics in feed induce prophages in swine fecal microbiomes
Heather K Allen et al. mBio. 2011.
Abstract
Antibiotics are a cost-effective tool for improving feed efficiency and preventing disease in agricultural animals, but the full scope of their collateral effects is not understood. Antibiotics have been shown to mediate gene transfer by inducing prophages in certain bacterial strains; therefore, one collateral effect could be prophage induction in the gut microbiome at large. Here we used metagenomics to evaluate the effect of two antibiotics in feed (carbadox and ASP250 [chlortetracycline, sulfamethazine, and penicillin]) on swine intestinal phage metagenomes (viromes). We also monitored the bacterial communities using 16S rRNA gene sequencing. ASP250, but not carbadox, caused significant population shifts in both the phage and bacterial communities. Antibiotic resistance genes, such as multidrug resistance efflux pumps, were identified in the viromes, but in-feed antibiotics caused no significant changes in their abundance. The abundance of phage integrase-encoding genes was significantly increased in the viromes of medicated swine over that in the viromes of nonmedicated swine, demonstrating the induction of prophages with antibiotic treatment. Phage-bacterium population dynamics were also examined. We observed a decrease in the relative abundance of Streptococcus bacteria (prey) when Streptococcus phages (predators) were abundant, supporting the "kill-the-winner" ecological model of population dynamics in the swine fecal microbiome. The data show that gut ecosystem dynamics are influenced by phages and that prophage induction is a collateral effect of in-feed antibiotics.
Importance: This study advances our knowledge of the collateral effects of in-feed antibiotics at a time in which the widespread use of "growth-promoting" antibiotics in agriculture is under scrutiny. Using comparative metagenomics, we show that prophages are induced by in-feed antibiotics in swine fecal microbiomes and that antibiotic resistance genes were detected in most viromes. This suggests that in-feed antibiotics are contributing to phage-mediated gene transfer, potentially of antibiotic resistance genes, in the swine gut. Additionally, the so-called "kill-the-winner" model of phage-bacterium population dynamics has been shown in aquatic ecosystems but met with conflicting evidence in gut ecosystems. The data support the idea that swine fecal Streptococcus bacteria and their phages follow the kill-the-winner model. Understanding the role of phages in gut microbial ecology is an essential component of the antibiotic resistance problem and of developing potential mitigation strategies.
Figures
FIG 1
Electron micrographs of virions isolated from swine feces. (A to D) Representative phages of the Myoviridae (A), Siphoviridae (B [no arrow] and C), and Podoviridae (B [arrow] and D) families. (E) An enveloped virus as seen in numerous fecal samples of young pigs. (F) Ten Siphoviridae (arrows) were visualized in a single field.
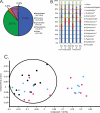
FIG 2
Community structure based on taxonomic inference of phages from swine feces. (A) Phage metagenomic sequence origins. “No hits” is the percentage of reads with no similar sequences in the database, and “unassigned or unclassified” is the percentage of reads with a database hit that has no associated taxonomic information. The values on the graph are the medians, and the values in parentheses are the ranges. (B) Genus-level phylogenetic origins of the phage-derived sequences from the ASP250 experiment. Phage taxa occurring at roughly <0.5% abundance were grouped as “Other phages.” Asterisks denote those of the Firmicutes phylum.
FIG 3
Community structure based on taxonomic inference of bacteria (16S rRNA sequences) from swine feces. (A) Phylum-level assignments of assignable 16S rRNA gene sequences from swine feces, averaged across all 86 individual samples. The values on the graph are the medians among treatment groups, and the values in parentheses are the ranges. (B) Average percent abundance of genus-level assignments of 16S rRNA gene sequences from the feces of six swine fed ASP250 and the corresponding nonmedicated animals. Values were normalized to the total number of assignments within a sample. Taxa occurring at roughly <0.3% abundance were grouped as “Others.” Asterisks denote those of the Firmicutes phylum. See Table S1 in the supplemental material for the values of the means and the standard errors. (C) Principal component analysis of OTU-based bacterial 16S rRNA gene sequence abundances in individual pig samples (P < 0.01, R = 0.43). The percent variance accounted for by each component is in parentheses. An ellipse is drawn around the data sets of pigs that did not receive ASP250. Black, day 0 (just prior to treatment); blue, day 8; pink, day 14. Circles, nonmedicated pigs; squares, medicated pigs.
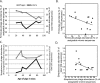
FIG 4
Population dynamics of bacteria and phages in swine fecal microbiomes. (A and C) Abundances of Firmicutes (A) and Streptococcus (C) bacteria and phages in the nonmedicated swine are plotted against time. (B and D) Regression analyses of the abundances of Firmicutes (B) and Streptococcus (D) phages against the respective bacterial abundances in all treatments and time points (_r_2 = 0.21 and 0.23, respectively; P < 0.1 for both). In all figures, the bacterial abundances are pooled data from six animals.
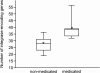
FIG 5
Box plot of integrase-encoding gene abundance in nonmedicated (n = 10) and medicated (n = 5) swine viromes (P < 0.01). Asterisks denote the means. The number of integrase-encoding genes was normalized by the total number of reads per virome.
Similar articles
- The In-Feed Antibiotic Carbadox Induces Phage Gene Transcription in the Swine Gut Microbiome.
Johnson TA, Looft T, Severin AJ, Bayles DO, Nasko DJ, Wommack KE, Howe A, Allen HK. Johnson TA, et al. mBio. 2017 Aug 8;8(4):e00709-17. doi: 10.1128/mBio.00709-17. mBio. 2017. PMID: 28790203 Free PMC article. - Prophages in Lactobacillus reuteri Are Associated with Fitness Trade-Offs but Can Increase Competitiveness in the Gut Ecosystem.
Oh JH, Lin XB, Zhang S, Tollenaar SL, Özçam M, Dunphy C, Walter J, van Pijkeren JP. Oh JH, et al. Appl Environ Microbiol. 2019 Dec 13;86(1):e01922-19. doi: 10.1128/AEM.01922-19. Print 2019 Dec 13. Appl Environ Microbiol. 2019. PMID: 31676478 Free PMC article. - Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations.
Looft T, Allen HK, Cantarel BL, Levine UY, Bayles DO, Alt DP, Henrissat B, Stanton TB. Looft T, et al. ISME J. 2014 Aug;8(8):1566-76. doi: 10.1038/ismej.2014.12. Epub 2014 Feb 13. ISME J. 2014. PMID: 24522263 Free PMC article. - The Human Gut Phage Community and Its Implications for Health and Disease.
Manrique P, Dills M, Young MJ. Manrique P, et al. Viruses. 2017 Jun 8;9(6):141. doi: 10.3390/v9060141. Viruses. 2017. PMID: 28594392 Free PMC article. Review. - Cryptic prophages as targets for drug development.
Wang X, Wood TK. Wang X, et al. Drug Resist Updat. 2016 Jul;27:30-8. doi: 10.1016/j.drup.2016.06.001. Epub 2016 Jun 6. Drug Resist Updat. 2016. PMID: 27449596 Review.
Cited by
- A composite bacteriophage alters colonization by an intestinal commensal bacterium.
Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. Duerkop BA, et al. Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17621-6. doi: 10.1073/pnas.1206136109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045666 Free PMC article. - Human oral viruses are personal, persistent and gender-consistent.
Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT. Abeles SR, et al. ISME J. 2014 Sep;8(9):1753-67. doi: 10.1038/ismej.2014.31. Epub 2014 Mar 20. ISME J. 2014. PMID: 24646696 Free PMC article. - Antimicrobial Effects on Swine Gastrointestinal Microbiota and Their Accompanying Antibiotic Resistome.
Zeineldin M, Aldridge B, Lowe J. Zeineldin M, et al. Front Microbiol. 2019 May 15;10:1035. doi: 10.3389/fmicb.2019.01035. eCollection 2019. Front Microbiol. 2019. PMID: 31156580 Free PMC article. Review. - Metagenomics and Other Omics Approaches to Bacterial Communities and Antimicrobial Resistance Assessment in Aquacultures.
Nogueira T, Botelho A. Nogueira T, et al. Antibiotics (Basel). 2021 Jun 28;10(7):787. doi: 10.3390/antibiotics10070787. Antibiotics (Basel). 2021. PMID: 34203511 Free PMC article. Review. - Estimating and comparing microbial diversity in the presence of sequencing errors.
Chiu CH, Chao A. Chiu CH, et al. PeerJ. 2016 Feb 1;4:e1634. doi: 10.7717/peerj.1634. eCollection 2016. PeerJ. 2016. PMID: 26855872 Free PMC article.
References
- Infectious Diseases Society of America 14 July 2010. The Infectious Diseases Society of America’s (IDSDA) statement on antibiotic resistance: promoting judicious use of medically important antibiotics in animal agriculture. Testimony before the House Committee on Energy and Commerce Subcommittee on Health. House Committee on Energy and Commerce, Washington, DC: http://democrats.energycommerce.house.gov/documents/20100714/Johnson.Tes...
- Center for Veterinary Medicine, Food and Drug Administration, US Department of Health and Human Services 2010. The judicious use of medically important antimicrobial drugs in food-producing animals. Draft guidance no. 209. Center for Veterinary Medicine, Food and Drug Administration, US Department of Health and Human Services, Rockville, MD
- US Government Accountability Office 2011. Antibiotic resistance: agencies have made limited progress addressing antibiotic use in animals. Report no. GAO-11-801. US Government Accountability Office, Washington, DC.
- Cromwell GL. 2002. Why and how antibiotics are used in swine production. Anim. Biotechnol. 13:7–27 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous