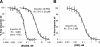Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5 - PubMed (original) (raw)
Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5
Ruth A Pumroy et al. J Biol Chem. 2012.
Abstract
The human genome encodes six isoforms of importin α that show greater than 60% sequence similarity and remarkable substrate specificity. The isoform importin α5 can bind phosphorylated cargos such as STAT1 and Epstein-Barr Virus Nuclear Antigen 1, as well as the influenza virus polymerase subunit PB2. In this work, we have studied the interaction of the nucleoporin Nup50 with importin α5. We show that the first 47 residues of Nup50 bind to the C terminus of importin α5 like a "clip," stabilizing the closed conformation of ARM 10. In vitro, Nup50 binds with high affinity either to empty importin α5 or to a preassembled complex of importin α5 bound to the C-terminal domain of the import cargo PB2, resulting in a trimeric complex. By contrast, PB2 can only bind with high affinity to importin α5 in the absence of Nup50. This suggests that Nup50 primary function may not be to actively displace the import cargo from importin α5 but rather to prevent cargo rebinding in preparation for recycling. This is the first evidence for a nucleoporin modulating the import reaction by directly altering the three-dimensional structure of an import adaptor.
Figures
FIGURE 1.
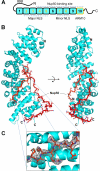
Structure of the ΔIBB-importin α5 bound to Nup50 (residues 1–47). A, schematic diagram of importin α5 domain structure that consists of an N-terminal IBB domain, an ARM core, and a C-terminal tail. The major and minor NLS-binding sites as well as the Nup50-binding region and ARM 10 are indicated. B, ribbon representation of the human ΔIBB-importin α5 (in cyan) bound to residues 1–47 of Nup50 (in red) shown in sticks. The nucleoporin wraps around the C terminus of importin α making contacts with ARMs 4–10. C, magnified view of a region of the Nup50 refined model (residues 1–7) superimposed to a final 2_F_o − _F_c electron density map (contoured at 1.1 σ above noise).
FIGURE 2.
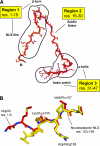
Extended structure of Nup50. A, regions of Nup50 responsible for interactions with importin α5. B, Nup50 occupies the minor NLS-binding site of importin α5: structural superimposition of residues 1–7 of Nup50 (in red) to residues 153–159 of nucleoplasmin NLS (in yellow) (Protein Data Bank 1EJY). The main determinants that bind to importin α are Lys3/Lys155, Arg4/Arg156, and Val5/Pro157.
FIGURE 3.
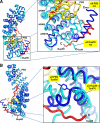
Flexibility of ARM 10 of importin α5. A, left panel, the structure of ΔIBB-importin α5 in complex with PB2 (Protein Data Bank 2JDQ) (in blue and yellow, respectively) is superimposed to that of ΔIBB-importin α5 bound to Nup50 (in cyan and red, respectively). Right panel, magnified view of ARM 8–10. Helix H3 in ARM 10 points in opposite directions in the two conformations of importin α5. B, left panel, superimposition of ΔIBB-importin α5 with PB2 and Nup50 as in A but rotated by 180°. Right panel, magnified view of Tyr476 that swings by 180° in response to Nup50 binding, thereby allowing stacking of ARM 10 helices H1–H3.
FIGURE 4.
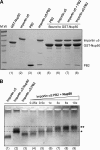
Nup50 and PB2 can simultaneously bind to importin α5. A, pulldown assay on glutathione-agarose beads coupled to GST-Nup50 (1–109) (lane 1). GST-Nup50 efficiently pulled down free ΔIBB-importin α5 (lane 6) but not PB2 (lane 5). Adding to the GST-Nup50 beads, either a preformed ΔIBB-importin α5-PB2 complex (lane 8) or free ΔIBB-importin α5 followed by an excess of PB2 (lane 7) results in formation of a trimeric complex. B, EMSA on native agarose gel. Free ΔIBB-importin α5 is in lane 1, and preformed complexes of ΔIBB-importin α5 bound to Nup50 or PB2 are in lanes 2 and 3, respectively. The addition of increasing quantities of Nup50 (from 0.25- to 10-fold molar excess) to a preformed ΔIBB-importin α5-PB2 complex leads to a trimeric species that is retarded as compared with ΔIBB-importin α5 bound to Nup50 (lanes 2) or PB2 (lane 3). Single and double asterisks indicate the position of the dimeric ΔIBB-importin α5-Nup50 and trimeric ΔIBB-importin α5-Nup50-PB2 complex, respectively. The difference in migration between these two species is also emphasized by dashed horizontal lines. M.W., molecular mass.
FIGURE 5.
Binding affinity of importin α5 for PB2 and Nup50 determined by flow fluorimetry. A, a titration of Nup50 was performed for a range of concentrations of free or PB2-bound ΔIBB-importin α5. The KD values were determined by fitting each set of data to a bimolecular equilibrium binding model (solid lines). Titrations of Nup50 against 100 p
m
empty ΔIBB-importin α5 gave a KD < 2 p
m
(left curve); titrations of Nup50 against 2 n
m
importin α5 prebound to an excess of PB2 resulted in a KD of 815 ± 29 p
m
(right curve). B, titrations of PB2 against 200 p
m
ΔIBB-importin α5 gave a KD of 53 ± 2 p
m
.
FIGURE 6.
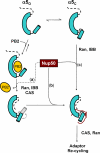
Model of involvement of Nup50 in importin α5 import cycle. Schematic diagram of the binding interactions of importin α5, Nup50, and PB2. Free importin α5 oscillates between an open (α5O) and closed (α5C) conformation of ARM 10; binding of PB2 or Nup50 to importin α5 stabilizes a given conformation. In all diagrams, the IBB domain of importin α5 is dashed, and for simplicity, importin β is not drawn.
Similar articles
- Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling.
Matsuura Y, Stewart M. Matsuura Y, et al. EMBO J. 2005 Nov 2;24(21):3681-9. doi: 10.1038/sj.emboj.7600843. Epub 2005 Oct 13. EMBO J. 2005. PMID: 16222336 Free PMC article. - Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5.
Nardozzi J, Wenta N, Yasuhara N, Vinkemeier U, Cingolani G. Nardozzi J, et al. J Mol Biol. 2010 Sep 10;402(1):83-100. doi: 10.1016/j.jmb.2010.07.013. Epub 2010 Jul 17. J Mol Biol. 2010. PMID: 20643137 Free PMC article. - Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import.
Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. Lindsay ME, et al. Cell. 2002 Aug 9;110(3):349-60. doi: 10.1016/s0092-8674(02)00836-x. Cell. 2002. PMID: 12176322 - Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import.
Ogawa Y, Miyamoto Y, Asally M, Oka M, Yasuda Y, Yoneda Y. Ogawa Y, et al. Mol Biol Cell. 2010 Feb 15;21(4):630-8. doi: 10.1091/mbc.e09-05-0374. Epub 2009 Dec 16. Mol Biol Cell. 2010. PMID: 20016008 Free PMC article. - Diversification of importin-α isoforms in cellular trafficking and disease states.
Pumroy RA, Cingolani G. Pumroy RA, et al. Biochem J. 2015 Feb 15;466(1):13-28. doi: 10.1042/BJ20141186. Biochem J. 2015. PMID: 25656054 Free PMC article. Review.
Cited by
- The Nup153-Nup50 protein interface and its role in nuclear import.
Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. Makise M, et al. J Biol Chem. 2012 Nov 9;287(46):38515-22. doi: 10.1074/jbc.M112.378893. Epub 2012 Sep 24. J Biol Chem. 2012. PMID: 23007389 Free PMC article. - Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein.
Nguyen NLT, Panté N. Nguyen NLT, et al. Cells. 2022 Sep 22;11(19):2957. doi: 10.3390/cells11192957. Cells. 2022. PMID: 36230922 Free PMC article. - Small terminase couples viral DNA binding to genome-packaging ATPase activity.
Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G. Roy A, et al. Structure. 2012 Aug 8;20(8):1403-13. doi: 10.1016/j.str.2012.05.014. Epub 2012 Jul 5. Structure. 2012. PMID: 22771211 Free PMC article. - Structure of Importin-α from a Filamentous Fungus in Complex with a Classical Nuclear Localization Signal.
Bernardes NE, Takeda AA, Dreyer TR, Freitas FZ, Bertolini MC, Fontes MR. Bernardes NE, et al. PLoS One. 2015 Jun 19;10(6):e0128687. doi: 10.1371/journal.pone.0128687. eCollection 2015. PLoS One. 2015. PMID: 26091498 Free PMC article. - Exploration of binary virus-host interactions using an infectious protein complementation assay.
Munier S, Rolland T, Diot C, Jacob Y, Naffakh N. Munier S, et al. Mol Cell Proteomics. 2013 Oct;12(10):2845-55. doi: 10.1074/mcp.M113.028688. Epub 2013 Jul 1. Mol Cell Proteomics. 2013. PMID: 23816991 Free PMC article.
References
- Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 - PubMed
- Mosammaparast N., Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 - PubMed
- Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous