Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides - PubMed (original) (raw)
Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides
David A Sela et al. Appl Environ Microbiol. 2012 Feb.
Abstract
Bifidobacterium longum subsp. infantis ATCC 15697 utilizes several small-mass neutral human milk oligosaccharides (HMOs), several of which are fucosylated. Whereas previous studies focused on endpoint consumption, a temporal glycan consumption profile revealed a time-dependent effect. Specifically, among preferred HMOs, tetraose was favored early in fermentation, with other oligosaccharides consumed slightly later. In order to utilize fucosylated oligosaccharides, ATCC 15697 possesses several fucosidases, implicating GH29 and GH95 α-L-fucosidases in a gene cluster dedicated to HMO metabolism. Evaluation of the biochemical kinetics demonstrated that ATCC 15697 expresses three fucosidases with a high turnover rate. Moreover, several ATCC 15697 fucosidases are active on the linkages inherent to the HMO molecule. Finally, the HMO cluster GH29 α-L-fucosidase possesses a crystal structure that is similar to previously characterized fucosidases.
Figures
Fig 1
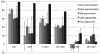
Temporal glycoprofile of abundant neutral HMO consumption by B. longum subsp. infantis ATCC 15697. HMO composition results are shown for a representative isomer signifying a characteristic oligosaccharide composition. All samples were analyzed at least in duplicate. LNT, lacto-_N_-tetraose-like composition; LNH, lacto-_N_-hexaose-like composition; F-LNH, fucosylated lacto-_N_-hexaose-like composition; DF-LNH, difucosylated lacto-_N_-hexaose-like composition; F-LNO, fucosylated lacto-_N_-octaose-like composition; DF-LNO, difucosylated lacto-_N_-hexaose-like composition.
Fig 2
Phylogenetic relationships of fucosidases encoded by select bacteria. Branch lengths are in the same units (number of amino acid substitutions per site) as those of the evolutionary distances used to construct the tree. The evolutionary history was inferred by the maximum likelihood method, followed by 100 bootstrapped replicates. The organism and loci are listed for those fucosidases found in glycoside hydrolase family 29 (A) and glycoside hydrolase family 95 (B).
Fig 3
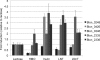
ATCC 15697 fucosidase gene expression during carbohydrate fermentation. Gene expression was calculated relative to levels when grown on lactose as the sole carbon source. Averages from three independent experiments are shown, and bars represent standard errors of the means.
Fig 4
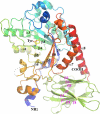
Ribbon drawing of the overall structure of Blon_2336. The N-terminal catalytic domain is shown by the various colors, from the blue at the N terminus to red at the C terminus. The β-strands and α-helices that form the (β/α)8 barrel are labeled in black. The C-terminal carbohydrate-binding domain is shown in lime green, with 9 β-strands labeled in magenta. A tyrosine from a possible short peptide at the active site of the catalytic domain (see description in the text) is drawn in stick format. A dashed curved line indicates a missing part in the final structural model (see the text). The N and C termini of Blon_2336 are also labeled.
Fig 5
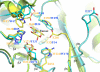
Structural alignment of the active sites of Blon_2336 and TM0306 in complex with fucose. Blon_2336 is shown in cyan, and TM0306 is shown in green-yellow. All active site residues and the fucose from the TM0306/fucose complex are drawn in stick format. The residues from Blon_2336 and TM0306 are labeled in blue and orange, respectively. Equivalent residues from two molecules are paired together. Some visible strands from the (β/α)8 barrel are labeled in black. The tyrosine shown in Fig. 4 is approximately at the position of the fucose in this figure, and it is not shown for the sake of clarity.
Similar articles
- Fucosylated Human Milk Oligosaccharide Foraging within the Species Bifidobacterium pseudocatenulatum Is Driven by Glycosyl Hydrolase Content and Specificity.
Shani G, Hoeflinger JL, Heiss BE, Masarweh CF, Larke JA, Jensen NM, Wickramasinghe S, Davis JC, Goonatilleke E, El-Hawiet A, Nguyen L, Klassen JS, Slupsky CM, Lebrilla CB, Mills DA. Shani G, et al. Appl Environ Microbiol. 2022 Jan 25;88(2):e0170721. doi: 10.1128/AEM.01707-21. Epub 2021 Nov 10. Appl Environ Microbiol. 2022. PMID: 34757822 Free PMC article. - Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense.
Bunesova V, Lacroix C, Schwab C. Bunesova V, et al. BMC Microbiol. 2016 Oct 26;16(1):248. doi: 10.1186/s12866-016-0867-4. BMC Microbiol. 2016. PMID: 27782805 Free PMC article. - Design of an α-L-transfucosidase for the synthesis of fucosylated HMOs.
Saumonneau A, Champion E, Peltier-Pain P, Molnar-Gabor D, Hendrickx J, Tran V, Hederos M, Dekany G, Tellier C. Saumonneau A, et al. Glycobiology. 2016 Mar;26(3):261-9. doi: 10.1093/glycob/cwv099. Epub 2015 Nov 17. Glycobiology. 2016. PMID: 26582607 - α-L-Fucosidases and their applications for the production of fucosylated human milk oligosaccharides.
Wan L, Zhu Y, Zhang W, Mu W. Wan L, et al. Appl Microbiol Biotechnol. 2020 Jul;104(13):5619-5631. doi: 10.1007/s00253-020-10635-7. Epub 2020 May 1. Appl Microbiol Biotechnol. 2020. PMID: 32356197 Review. - Enzymatic transfucosylation for synthesis of human milk oligosaccharides.
Zeuner B, Meyer AS. Zeuner B, et al. Carbohydr Res. 2020 Jul;493:108029. doi: 10.1016/j.carres.2020.108029. Epub 2020 May 8. Carbohydr Res. 2020. PMID: 32445980 Review.
Cited by
- Regulatory mechanism of cysteine-dependent methionine biosynthesis in Bifidobacterium longum: insights into sulfur metabolism in gut microbiota.
Kim YT, Kwon JG, O'Sullivan DJ, Lee JH. Kim YT, et al. Gut Microbes. 2024 Jan-Dec;16(1):2419565. doi: 10.1080/19490976.2024.2419565. Epub 2024 Oct 28. Gut Microbes. 2024. PMID: 39468828 Free PMC article. - Impact of Bifidobacterium longum Subspecies infantis on Pediatric Gut Health and Nutrition: Current Evidence and Future Directions.
Dargenio VN, Cristofori F, Brindicci VF, Schettini F, Dargenio C, Castellaneta SP, Iannone A, Francavilla R. Dargenio VN, et al. Nutrients. 2024 Oct 16;16(20):3510. doi: 10.3390/nu16203510. Nutrients. 2024. PMID: 39458503 Free PMC article. Review. - Determining the metabolic fate of human milk oligosaccharides: it may just be more complex than you think?
Jackson PPJ, Wijeyesekera A, Rastall RA. Jackson PPJ, et al. Gut Microbiome (Camb). 2022 Sep 7;3:e9. doi: 10.1017/gmb.2022.8. eCollection 2022. Gut Microbiome (Camb). 2022. PMID: 39295778 Free PMC article. Review. - Modifiable and Non-Modifiable Factors That Affect Human Milk Oligosaccharides Composition.
Konieczna M, Koryszewska-Bagińska A, Bzikowska-Jura A, Chmielewska-Jeznach M, Jarzynka S, Olędzka G. Konieczna M, et al. Nutrients. 2024 Aug 28;16(17):2887. doi: 10.3390/nu16172887. Nutrients. 2024. PMID: 39275203 Free PMC article. Review. - Integrative genomic reconstruction of carbohydrate utilization networks in bifidobacteria: global trends, local variability, and dietary adaptation.
Arzamasov AA, Rodionov DA, Hibberd MC, Guruge JL, Kazanov MD, Leyn SA, Kent JE, Sejane K, Bode L, Barratt MJ, Gordon JI, Osterman AL. Arzamasov AA, et al. bioRxiv [Preprint]. 2024 Jul 7:2024.07.06.602360. doi: 10.1101/2024.07.06.602360. bioRxiv. 2024. PMID: 39005317 Free PMC article. Preprint.
References
- Ashida H, et al. 2009. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017 - PubMed
- Cohen SX, et al. 2004. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60:2222–2229 - PubMed
- Collaborative Computation Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 - PubMed
- Emsley P, Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM074942/GM/NIGMS NIH HHS/United States
- T32-GM08799/GM/NIGMS NIH HHS/United States
- R01 HD059127/HD/NICHD NIH HHS/United States
- U54 GM074942/GM/NIGMS NIH HHS/United States
- R01HD061923/HD/NICHD NIH HHS/United States
- R01HD059127/HD/NICHD NIH HHS/United States
- R01 HD061923/HD/NICHD NIH HHS/United States
- T32 GM008799/GM/NIGMS NIH HHS/United States
- R01 HD065122/HD/NICHD NIH HHS/United States
- R01HD065122/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources