Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall - PubMed (original) (raw)
Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall
Masayuki Hashimoto et al. J Bacteriol. 2012 Feb.
Abstract
Bacterial peptidoglycan acts as an exoskeleton to protect the bacterial cell. Although peptidoglycan biosynthesis by penicillin-binding proteins is well studied, few studies have described peptidoglycan disassembly, which is necessary for a dynamic structure that allows cell growth. In Bacillus subtilis, more than 35 genes encoding cell wall lytic enzymes have been identified; however, only two D,L-endopeptidases (lytE and cwlO) are involved in cell proliferation. In this study, we demonstrated that the D,L-endopeptidase activity at the lateral cell wall is essential for cell proliferation. Inactivation of LytE and CwlO by point mutation of the catalytic residues caused cell growth defects. However, the forced expression of LytF or CwlS, which are paralogs of LytE, did not suppress lytE cwlO synthetic lethality. Subcellular localization studies of these D,L-endopeptidases showed LytF and CwlS at the septa and poles, CwlO at the cylindrical part of the cell, and LytE at the septa and poles as well as the cylindrical part. Furthermore, construction of N-terminal and C-terminal domain-swapped enzymes of LytE, LytF, CwlS, and CwlO revealed that localization was dependent on the N-terminal domains. Only the chimeric proteins that were enzymatically active and localized to the sidewall were able to suppress the synthetic lethality, suggesting that the lack of D,L-endopeptidase activity at the cylindrical part of the cell leads to a growth defect. The functions of LytE and CwlO in cell morphogenesis were discussed.
Figures
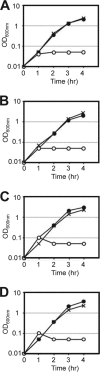
Fig 1
d
,
l
-Endopeptidase activity of LytE and CwlO is important for cell proliferation, and LytF or CwlS induction could not suppress lytE cwlO synthetic lethality. Strains were precultured with the appropriate inducer until late exponential phase (OD600 = 2.0). An aliquot of each culture was washed and inoculated into fresh medium with or without the inducer to an OD600 of 0.01. The × symbol in panels A to D indicates the wild-type 168 strain. (A) Growth of OH005 [lytE(C247S)-6×FLAG P_xyl-cwlO; open circles] and OH004 (lytE_-6×FLAG P_xyl-cwlO; closed circles). Xylose (1%) was added to the preculture, but CwlO expression was not induced by xylose in the main culture. (B) Growth of OH007 [cwlO(C377S)-6×FLAG P_spac-lytE; open circles] and OH006 (cwlO_-6×FLAG P_spac-lytE; closed circles). IPTG (1 mM) was added to the preculture, but LytE expression was not induced by IPTG in the main culture. (C) Growth of OH009 (Δ_lytE P_xyl-cwlO P_spac-lytF_). The strain was cultured with 1 mM IPTG to induce LytF expression and with 1% xylose to induce CwlO induction (closed circles) or without xylose (open circles). (D) Growth of OH012 (Δ_lytE_ P_xyl-cwlO_ P_spac-cwlS_). The strain was cultured with 1 mM IPTG to induce CwlS expression and with 1% xylose to induce CwlO expression (closed circles) or without xylose (open circles).
Fig 2
Subcellular localization of full-length
d
,
l
-endopeptidases and their N-terminal domains. Phase-contrast and immunofluorescence microscopy analysis of FLAG-tagged proteins. The OD600 values at the sampling times were 0.1 for LytE and CwlO and their N-terminal domains (CWBLytE and NTDCwlO, respectively), 0.6 for LytF and its N-terminal domain (CWBLytF), and 2.0 for CwlS and its N-terminal domain (CWBCwlS). (A) WECLytE6FL (LytE-6×FLAG); (B) OH015 (CWBLytE-6×FLAG); (C) WECLytF6FL (LytF-6×FLAG); (D) OH014 (CWBLytF-6×FLAG); (E) WECS6FL (CwlS-6×FLAG); (F) OH016 (CWBCwlS-6×FLAG); (G) WECO6FL (CwlO-6×FLAG); (H) OH013 (overexpressed CwlO-6×FLAG); and (I) OH018 (overexpressed NTDCwlO-6×FLAG). Bars = 5 μm.
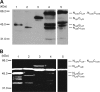
Fig 3
Expression and activity of domain-swapped
d
,
l
-endopeptidases. Strains were exposed to 1% xylose or 1 mM IPTG for 2 h to induce P_xyl-cwlO_ and P_spac-lytE_ expression, respectively. Lanes: 1, OH019 (NLytECLytF P_xyl-cwlO_, 41 kDa); 2, OH020 (NLytECCwlS P_xyl-cwlO_, 40 kDa); 3, OH022 (NLytFCLytE P_xyl-cwlO_, 53 kDa); 4, OH023 (NCwlOCLytF P_spac-lytE_, 55 kDa); and 5, OH024 (NCwlOCCwlS P_spac-lytE_, 56 kDa). (A) Domain-swapped
d
,
l
-endopeptidases were evaluated by Western blot analysis with an anti-FLAG antibody. Degraded products of the chimeric enzymes appear in lanes 4 and 5. (B) Zymography of the chimeric enzymes using B. subtilis cell wall as a substrate. Asterisks indicate clear zones produced by the chimeric enzymes.
Fig 4
Subcellular localization of domain-swapped
d
,
l
-endopeptidases and suppression of the lytE cwlO synthetic lethality by these proteins. For microscopic imaging, OH019 (lytE::NLytECLytF P_xyl-cwlO_), OH020 (lytE::NLytECCwlS P_xyl-cwlO_), and OH022 (lytE::NLytFCLytE P_xyl-cwlO_) were cultured with 1% xylose to induce CwlO, and OH023 (cwlO::NCwlOCLytF P_spac-lytE_) and OH024 (cwlO::NCwlOCCwlS P_spac-lytE_) were cultured with 1 mM IPTG to induce LytE. For suppression assays, the strains were grown under the same conditions as those described in Fig. 1. They were cultured with xylose (closed circles) or without xylose (open circles) for P_xyl-cwlO_ and IPTG for P_spac-lytE_. The × symbol indicates the wild-type 168 strain. Bars = 5 μm.
Similar articles
- The WalR-WalK Signaling Pathway Modulates the Activities of both CwlO and LytE through Control of the Peptidoglycan Deacetylase PdaC in Bacillus subtilis.
Dobihal GS, Flores-Kim J, Roney IJ, Wang X, Rudner DZ. Dobihal GS, et al. J Bacteriol. 2022 Feb 15;204(2):e0053321. doi: 10.1128/JB.00533-21. Epub 2021 Dec 6. J Bacteriol. 2022. PMID: 34871030 Free PMC article. - Disruption of the cell wall lytic enzyme CwlO affects the amount and molecular size of poly-γ-glutamic acid produced by Bacillus subtilis (natto).
Mitsui N, Murasawa H, Sekiguchi J. Mitsui N, et al. J Gen Appl Microbiol. 2011;57(1):35-43. doi: 10.2323/jgam.57.35. J Gen Appl Microbiol. 2011. PMID: 21478646 - Digestion of peptidoglycan near the cross-link is necessary for the growth of Bacillus subtilis.
Hashimoto M, Matsushima H, Suparthana IP, Ogasawara H, Yamamoto H, Teng C, Sekiguchi J. Hashimoto M, et al. Microbiology (Reading). 2018 Mar;164(3):299-307. doi: 10.1099/mic.0.000614. Epub 2018 Jan 25. Microbiology (Reading). 2018. PMID: 29458657 - Autolysins of Bacillus subtilis: multiple enzymes with multiple functions.
Smith TJ, Blackman SA, Foster SJ. Smith TJ, et al. Microbiology (Reading). 2000 Feb;146 ( Pt 2):249-262. doi: 10.1099/00221287-146-2-249. Microbiology (Reading). 2000. PMID: 10708363 Review. No abstract available. - Lytic transglycosylases: bacterial space-making autolysins.
Scheurwater E, Reid CW, Clarke AJ. Scheurwater E, et al. Int J Biochem Cell Biol. 2008;40(4):586-91. doi: 10.1016/j.biocel.2007.03.018. Epub 2007 Mar 30. Int J Biochem Cell Biol. 2008. PMID: 17468031 Review.
Cited by
- Comprehensive double-mutant analysis of the Bacillus subtilis envelope using double-CRISPRi.
Koo BM, Todor H, Sun J, van Gestel J, Hawkins JS, Hearne CC, Banta AB, Huang KC, Peters JM, Gross CA. Koo BM, et al. bioRxiv [Preprint]. 2024 Aug 16:2024.08.14.608006. doi: 10.1101/2024.08.14.608006. bioRxiv. 2024. PMID: 39185233 Free PMC article. Preprint. - Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria.
Yunck R, Cho H, Bernhardt TG. Yunck R, et al. Mol Microbiol. 2016 Feb;99(4):700-18. doi: 10.1111/mmi.13258. Epub 2015 Nov 19. Mol Microbiol. 2016. PMID: 26507882 Free PMC article. - Evidence that glycopolymer transferases promote peptidoglycan hydrolysis in Bacillus subtilis.
Cohen JD. Cohen JD. bioRxiv [Preprint]. 2025 Mar 11:2025.02.26.640348. doi: 10.1101/2025.02.26.640348. bioRxiv. 2025. PMID: 40060662 Free PMC article. Preprint. - Peptidoglycan hydrolase of an unusual cross-link cleavage specificity contributes to bacterial cell wall synthesis.
Chodisetti PK, Reddy M. Chodisetti PK, et al. Proc Natl Acad Sci U S A. 2019 Apr 16;116(16):7825-7830. doi: 10.1073/pnas.1816893116. Epub 2019 Apr 2. Proc Natl Acad Sci U S A. 2019. PMID: 30940749 Free PMC article. - Recent advances in understanding how rod-like bacteria stably maintain their cell shapes.
van Teeffelen S, Renner LD. van Teeffelen S, et al. F1000Res. 2018 Feb 28;7:241. doi: 10.12688/f1000research.12663.1. eCollection 2018. F1000Res. 2018. PMID: 29560261 Free PMC article. Review.
References
- Aramini JM, et al. 2008. Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47:9715–9717 - PubMed
- Bisicchia P, et al. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180–200 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases