BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata - PubMed (original) (raw)
. 2012 Jan;40(Database issue):D57-63.
doi: 10.1093/nar/gkr1163. Epub 2011 Dec 1.
Karen Clark, Robert Gevorgyan, Vyacheslav Gorelenkov, Eugene Gribov, Ilene Karsch-Mizrachi, Michael Kimelman, Kim D Pruitt, Sergei Resenchuk, Tatiana Tatusova, Eugene Yaschenko, James Ostell
Affiliations
- PMID: 22139929
- PMCID: PMC3245069
- DOI: 10.1093/nar/gkr1163
BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata
Tanya Barrett et al. Nucleic Acids Res. 2012 Jan.
Abstract
As the volume and complexity of data sets archived at NCBI grow rapidly, so does the need to gather and organize the associated metadata. Although metadata has been collected for some archival databases, previously, there was no centralized approach at NCBI for collecting this information and using it across databases. The BioProject database was recently established to facilitate organization and classification of project data submitted to NCBI, EBI and DDBJ databases. It captures descriptive information about research projects that result in high volume submissions to archival databases, ties together related data across multiple archives and serves as a central portal by which to inform users of data availability. Concomitantly, the BioSample database is being developed to capture descriptive information about the biological samples investigated in projects. BioProject and BioSample records link to corresponding data stored in archival repositories. Submissions are supported by a web-based Submission Portal that guides users through a series of forms for input of rich metadata describing their projects and samples. Together, these databases offer improved ways for users to query, locate, integrate and interpret the masses of data held in NCBI's archival repositories. The BioProject and BioSample databases are available at http://www.ncbi.nlm.nih.gov/bioproject and http://www.ncbi.nlm.nih.gov/biosample, respectively.
Figures
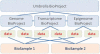
Figure 1.
Schematic depicting how BioProject, BioSample and data objects can be organized and linked. This example is composed of one umbrella project that encompasses three subprojects, each of which generated data derived from two BioSample records. Users can query either the BioProject or the BioSample database to retrieve the relevant records, and then navigate through links to the corresponding experimental data which continue to be stored in NCBI's primary data archives, including GenBank, SRA, dbGaP and GEO. This schematic depicts direct links that can be applied between objects; it does not depict links to corresponding records in other NCBI databases, including PubMed, Gene, Genome and Taxonomy.
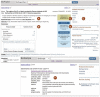
Figure 2.
Screenshot of a Genome Sequencing project that is a component of an umbrella project that encompasses data generated from an E. coli pathogen outbreak (upper panel) (17) and a corresponding sample record (lower panel). The records display the project title, summary, data type, locus_tag prefix and various project attributes including the scope and capture method (A). The Project Data section (B) lists the availability of corresponding sequence and assembly data in the Nucleotide and SRA databases where the data can be downloaded. Navigation panels assist users to link to Genome-level resources for that organism (C), or to ‘Navigate Up’ to the parent umbrella project, or to ‘Navigate across’ to sibling projects that are part of that umbrella project, as well as any additional projects related by organism (D). The ‘Related Information’ panel (E) contains full list of linkages for that record; clicking the BioSample link directs the user to the sample record shown in the lower panel, which lists the attributes that were collected for that sample including the collection date, isolation source, country and strain and serovar (F).
Similar articles
- "METAGENOTE: a simplified web platform for metadata annotation of genomic samples and streamlined submission to NCBI's sequence read archive".
Quiñones M, Liou DT, Shyu C, Kim W, Vujkovic-Cvijin I, Belkaid Y, Hurt DE. Quiñones M, et al. BMC Bioinformatics. 2020 Sep 3;21(1):378. doi: 10.1186/s12859-020-03694-0. BMC Bioinformatics. 2020. PMID: 32883210 Free PMC article. - BioSamples database: an updated sample metadata hub.
Courtot M, Cherubin L, Faulconbridge A, Vaughan D, Green M, Richardson D, Harrison P, Whetzel PL, Parkinson H, Burdett T. Courtot M, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D1172-D1178. doi: 10.1093/nar/gky1061. Nucleic Acids Res. 2019. PMID: 30407529 Free PMC article. - The CAIRR Pipeline for Submitting Standards-Compliant B and T Cell Receptor Repertoire Sequencing Studies to the National Center for Biotechnology Information Repositories.
Bukhari SAC, O'Connor MJ, Martínez-Romero M, Egyedi AL, Willrett D, Graybeal J, Musen MA, Rubelt F, Cheung KH, Kleinstein SH. Bukhari SAC, et al. Front Immunol. 2018 Aug 16;9:1877. doi: 10.3389/fimmu.2018.01877. eCollection 2018. Front Immunol. 2018. PMID: 30166985 Free PMC article. - Gene expression omnibus: microarray data storage, submission, retrieval, and analysis.
Barrett T, Edgar R. Barrett T, et al. Methods Enzymol. 2006;411:352-69. doi: 10.1016/S0076-6879(06)11019-8. Methods Enzymol. 2006. PMID: 16939800 Free PMC article. Review.
Cited by
- Toward Accurate and Quantitative Comparative Metagenomics.
Nayfach S, Pollard KS. Nayfach S, et al. Cell. 2016 Aug 25;166(5):1103-1116. doi: 10.1016/j.cell.2016.08.007. Cell. 2016. PMID: 27565341 Free PMC article. Review. - MarineMetagenomeDB: a public repository for curated and standardized metadata for marine metagenomes.
Nata'ala MK, Avila Santos AP, Coelho Kasmanas J, Bartholomäus A, Saraiva JP, Godinho Silva S, Keller-Costa T, Costa R, Gomes NCM, Ponce de Leon Ferreira de Carvalho AC, Stadler PF, Sipoli Sanches D, Nunes da Rocha U. Nata'ala MK, et al. Environ Microbiome. 2022 Nov 18;17(1):57. doi: 10.1186/s40793-022-00449-7. Environ Microbiome. 2022. PMID: 36401317 Free PMC article. - BioPS: System for screening and assessment of biofuel-production potential of cyanobacteria.
Motwalli O, Essack M, Salhi A, Hanks J, Mijakovic I, Bajic VB. Motwalli O, et al. PLoS One. 2018 Aug 10;13(8):e0202002. doi: 10.1371/journal.pone.0202002. eCollection 2018. PLoS One. 2018. PMID: 30096176 Free PMC article. - PDX Finder: A portal for patient-derived tumor xenograft model discovery.
Conte N, Mason JC, Halmagyi C, Neuhauser S, Mosaku A, Yordanova G, Chatzipli A, Begley DA, Krupke DM, Parkinson H, Meehan TF, Bult CC. Conte N, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D1073-D1079. doi: 10.1093/nar/gky984. Nucleic Acids Res. 2019. PMID: 30535239 Free PMC article. - Gypsy moth genome provides insights into flight capability and virus-host interactions.
Zhang J, Cong Q, Rex EA, Hallwachs W, Janzen DH, Grishin NV, Gammon DB. Zhang J, et al. Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1669-1678. doi: 10.1073/pnas.1818283116. Epub 2019 Jan 14. Proc Natl Acad Sci U S A. 2019. PMID: 30642971 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials