Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways - PubMed (original) (raw)
Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways
Neerav Shukla et al. Clin Cancer Res. 2012.
Abstract
Purpose: In contrast to the numerous broad screens for oncogene mutations in adult cancers, few such screens have been conducted in pediatric solid tumors. To identify novel mutations and potential therapeutic targets in pediatric cancers, we conducted a high-throughput Sequenom-based analysis in large sets of several major pediatric solid cancers, including neuroblastoma, Ewing sarcoma, rhabdomyosarcoma (RMS), and desmoplastic small round cell tumor (DSRCT).
Experimental design: We designed a highly multiplexed Sequenom-based assay to interrogate 275 recurrent mutations across 29 genes. Genomic DNA was extracted from 192 neuroblastoma, 75 Ewing sarcoma, 89 RMS, and 24 DSRCT samples. All mutations were verified by Sanger sequencing.
Results: Mutations were identified in 13% of neuroblastoma samples, 4% of Ewing sarcoma samples, 21.1% of RMS samples, and no DSRCT samples. ALK mutations were present in 10.4% of neuroblastoma samples. The remainder of neuroblastoma mutations involved the BRAF, RAS, and MAP2K1 genes and were absent in samples harboring ALK mutations. Mutations were more common in embryonal RMS (ERMS) samples (28.3%) than alveolar RMS (3.5%). In addition to previously identified RAS and FGFR4 mutations, we report for the first time PIK3CA and CTNNB1 (β-catenin) mutations in 5% and 3.3% of ERMS, respectively.
Conclusions: In ERMS, Ewing sarcoma, and neuroblastoma, we identified novel occurrences of several oncogene mutations recognized as drivers in other cancers. Overall, neuroblastoma and ERMS contain significant subsets of cases with nonoverlapping mutated genes in growth signaling pathways. Tumor profiling can identify a subset of pediatric solid tumor patients as candidates for kinase inhibitors or RAS-targeted therapies.
Figures
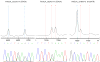
Figure 1. PIK3CA mutations in rhabdomyosarcoma
3 mutations were identified by Sequenom analysis and verified by direct sequencing. E542K and E545K mutations involve the helical domain of PIK3CA. The H1047R mutation involves the kinase domain of PIK3CA.
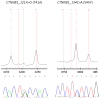
Figure 2. CTNNB1 mutations in rhabdomyosarcoma
2 mutations were identified by Sequenom analysis and verified by direct sequencing.
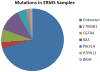
Figure 3. Oncogenic mutations identified in ERMS samples
Mutations in one of 6 known cancer genes were identified in approximately 28% of all ERMS samples tested.
Similar articles
- Frequent HRAS Mutations in Malignant Ectomesenchymoma: Overlapping Genetic Abnormalities With Embryonal Rhabdomyosarcoma.
Huang SC, Alaggio R, Sung YS, Chen CL, Zhang L, Kao YC, Agaram NP, Wexler LH, Antonescu CR. Huang SC, et al. Am J Surg Pathol. 2016 Jul;40(7):876-85. doi: 10.1097/PAS.0000000000000612. Am J Surg Pathol. 2016. PMID: 26872011 Free PMC article. - Comprehensive Molecular Profiling of Desmoplastic Small Round Cell Tumor.
Slotkin EK, Bowman AS, Levine MF, Dela Cruz F, Coutinho DF, Sanchez GI, Rosales N, Modak S, Tap WD, Gounder MM, Thornton KA, Bouvier N, You D, Gundem G, Gerstle JT, Heaton TE, LaQuaglia MP, Wexler LH, Meyers PA, Kung AL, Papaemmanuil E, Zehir A, Ladanyi M, Shukla N. Slotkin EK, et al. Mol Cancer Res. 2021 Jul;19(7):1146-1155. doi: 10.1158/1541-7786.MCR-20-0722. Epub 2021 Mar 22. Mol Cancer Res. 2021. PMID: 33753552 Free PMC article. - Anaplastic lymphoma kinase gene expression in small round cell tumors of childhood--a comparative ımmunohistochemical study.
Karakuş E, Emir S, Kaçar A, Karakuş R, Demir HA, Özyörük D. Karakuş E, et al. Ann Diagn Pathol. 2015 Aug;19(4):239-42. doi: 10.1016/j.anndiagpath.2015.04.007. Epub 2015 May 12. Ann Diagn Pathol. 2015. PMID: 26005112 - The role of anaplastic lymphoma kinase in pediatric cancers.
Takita J. Takita J. Cancer Sci. 2017 Oct;108(10):1913-1920. doi: 10.1111/cas.13333. Epub 2017 Aug 24. Cancer Sci. 2017. PMID: 28756644 Free PMC article. Review. - Phenotype and immunophenotype of the most common pediatric tumors.
Picarsic J, Reyes-Múgica M. Picarsic J, et al. Appl Immunohistochem Mol Morphol. 2015 May-Jun;23(5):313-26. doi: 10.1097/PAI.0000000000000068. Appl Immunohistochem Mol Morphol. 2015. PMID: 25390357 Review.
Cited by
- Sequencing Overview of Ewing Sarcoma: A Journey across Genomic, Epigenomic and Transcriptomic Landscapes.
Sand LG, Szuhai K, Hogendoorn PC. Sand LG, et al. Int J Mol Sci. 2015 Jul 16;16(7):16176-215. doi: 10.3390/ijms160716176. Int J Mol Sci. 2015. PMID: 26193259 Free PMC article. Review. - Strategies to inhibit FGFR4 V550L-driven rhabdomyosarcoma.
Fiorito E, Szybowska P, Haugsten EM, Kostas M, Øy GF, Wiedlocha A, Singh S, Nakken S, Mælandsmo GM, Fletcher JA, Meza-Zepeda LA, Wesche J. Fiorito E, et al. Br J Cancer. 2022 Nov;127(11):1939-1953. doi: 10.1038/s41416-022-01973-6. Epub 2022 Sep 12. Br J Cancer. 2022. PMID: 36097178 Free PMC article. - Emerging treatment options for the treatment of neuroblastoma: potential role of perifosine.
Sun W, Modak S. Sun W, et al. Onco Targets Ther. 2012;5:21-9. doi: 10.2147/OTT.S14578. Epub 2012 Mar 2. Onco Targets Ther. 2012. PMID: 22419878 Free PMC article. - Role of GOLPH3 and TPX2 in Neuroblastoma DNA Damage Response and Cell Resistance to Chemotherapy.
Ognibene M, Podestà M, Garaventa A, Pezzolo A. Ognibene M, et al. Int J Mol Sci. 2019 Sep 25;20(19):4764. doi: 10.3390/ijms20194764. Int J Mol Sci. 2019. PMID: 31557970 Free PMC article. - Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors.
Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, Tolman C, Hurd L, Liao H, Zhang S, Bogen D, Brohl AS, Sindiri S, Catchpoole D, Badgett T, Getz G, Mora J, Anderson JR, Skapek SX, Barr FG, Meyerson M, Hawkins DS, Khan J. Shern JF, et al. Cancer Discov. 2014 Feb;4(2):216-31. doi: 10.1158/2159-8290.CD-13-0639. Epub 2014 Jan 23. Cancer Discov. 2014. PMID: 24436047 Free PMC article.
References
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. - PubMed
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–9. - PubMed
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous