Mitochondrial dysfunction in ataxia-telangiectasia - PubMed (original) (raw)
Mitochondrial dysfunction in ataxia-telangiectasia
Yasmine A Valentin-Vega et al. Blood. 2012.
Abstract
Ataxia-telangiectasia mutated (ATM) plays a central role in DNA damage responses, and its loss leads to development of T-cell malignancies. Here, we show that ATM loss also leads to intrinsic mitochondrial abnormalities in thymocytes, including elevated reactive oxygen species, increased aberrant mitochondria, high cellular respiratory capacity, and decreased mitophagy. A fraction of ATM protein is localized in mitochondria, and it is rapidly activated by mitochondrial dysfunction. Unexpectedly, allelic loss of the autophagy regulator Beclin-1 significantly delayed tumor development in ATM-null mice. This effect was not associated with rescue of DNA damage signaling but rather with a significant reversal of the mitochondrial abnormalities. These data support a model in which ATM plays direct roles in modulating mitochondrial homeostasis and suggest that mitochondrial dysfunction and associated increases in mitochondrial reactive oxygen species contribute to the cancer-prone phenotype observed in organisms lacking ATM. Thus, ataxia-telangiectasia should be considered, at least in part, as a mitochondrial disease.
Figures
Figure 1
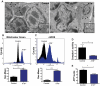
Mitochondrial dysfunction in _ATM_-null thymocytes. (A) Ultrastructural abnormalities in _ATM_-deficient thymic cells. Representative pictures taken by transmission electron microscopy in young mice from wild-type or _ATM_−/− mice. _ATM_−/− thymic tissue showed a number of mitochondria with disorganized structure and swollen appearance. Arrowheads denote mitochondria. Magnification, × 6000. Boxes show higher magnifications of the mitochondria. (B-C) Increased mitochondrial mass and mROS in _ATM_−/− thymocytes. Freshly isolated thymic cells were stained with 200nM MitoTracker Green (MTG) probe (B) or 5μM superoxide indicator MitoSOX (C) and analyzed by flow cytometry. Representative histograms of MTG and MitoSOX fluorescence intensity per genotype are illustrated. Graph shows the averaged mean intensity for each genotype (n ≥ 3/group; Student t test: **P ≤ .001, ***P ≤ .0001). (D-E) _ATM_-deficient thymocytes display reduced activity of complex I of the ETC and diminished levels of cellular ATP. (D) Total protein was extracted from freshly isolated viable thymocytes of 5- to 8-week-old mice of the indicated genotypes. The activity of complex I of the ETC was analyzed using equal amount of protein per sample (Student t test: *P ≤ .02; n ≥ 3/genotype). (E) _ATM_−/− thymocytes display reductions in cellular ATP levels. Total cellular ATP levels were measured in equal number of freshly isolated viable thymic cells isolated from 5- to 8-week-old mice of the indicated genotypes. Shown are the relative ATP levels for each cohort (Student t test: *P ≤ .02; n ≥ 3/genotype).
Figure 2
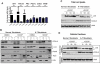
ATM deficiency is associated with abnormal mitophagy, not increased mitochondrial biogenesis. (A) Expression of genes involved in mitochondrial biogenesis is intact in _ATM_-deficient and wild-type thymocytes. Nrf1, _PGC1_β, PRC, _PGC1_α, SOD2, and TFAM mRNAs were measured by reverse transcription real-time PCR analyses. mRNA levels were normalized using Actin mRNA as an internal control. N.S. indicates not statistically significant. (B-D) Accumulation of Parkin and impaired mitophagy in A-T human fibroblasts. (B) Parkin levels are elevated in mitochondria of A-T fibroblasts without induction of mitochondrial damage by CCCP. Normal or A-T human fibroblasts were treated with DMSO or 50μM CCCP for 4 hours, and equal number of cells per sample was submitted to fractionation into mitochondrial (M), cytoplasmic (C), and nuclear (N) subregions. Equal volume of the fractions was analyzed by Western blot for the expression of Parkin, COX-IV (mitochondrial marker), Topoisomerase I (Topo; nuclear marker), LC3 (autophagic marker), and lactate dehydrogenase (LDH; cytoplasmic marker). We also analyzed 20 μg of total (T) protein. The small amount of LC3 present at the nuclear fraction after CCCP treatment is probably because of a residual mitochondrial material that fractionated with the nuclear compartment because CCCP could potentially induce damage to the mitochondrial membrane and thus make these organelles more leaky. (C) Normal or A-T human fibroblasts were treated with DMSO or 50μM CCCP for 3, 16, or 24 hours, and proteins were isolated followed by immunoblotting against COX-IV, LC3 (autophagic marker), and Actin (loading control) in total cell lysate. (D) Normal or A-T human fibroblasts were treated with CCCP as in panel C followed by cellular fractionation as in panel B, and mitochondrial protein fractions were analyzed by immunoblotting against COX-IV and LC3. Actin levels in the cytoplasmic fractions of these samples also were analyzed to verify equal loading based on cell number.
Figure 3
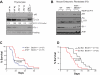
Beclin-1 heterozygosity reverts the induction of macroautophagy and reduced levels of the oxidative sensor DJ-1 in _ATM_-null cells but delays tumorigenesis in _ATM_-null mice. (A) Allelic loss of Beclin-1 reverses the increases in autophagic signaling and the reduced levels of DJ-1 manifest in _ATM_−/− thymocytes. Proteins from freshly isolated thymocytes were analyzed by SDS-PAGE immunoblotting for detection of the autophagic markers p62 and LC3 and the oxidative stress sensor DJ-1. Band intensities of the cytoplasmic (-I) and autophagosome-associated (-II) forms of LC3 were determined using the ImageJ processing program, and the ratio of the LC3-I/LC3-II forms was calculated for each sample. (B) Allelic loss of Beclin-1 reverses the increased autophagic response and the reductions in DJ-1 protein levels observed in early passage _ATM_−/− MEFs. P2 MEFs were treated with either DMSO or 50μM chloroquine (CQ; Sigma-Aldrich) for 4 hours, and total proteins were analyzed for the autophagic markers p62 and LC3, or for DJ-1. Transient treatment with CQ did not significantly alter p62 expression but affected LC3-I/II conversion in MEFs. (C-D) Allelic loss of Beclin-1 delays death of _ATM_−/− mice (C), while accelerating onset of B-cell lymphoma in Eμ_-Myc_–transgenic mice (D). Kaplan-Meier survival plots of mice with desired genotypes are shown. P values were obtained by the log-rank (Mantel-Cox) test. The nonsurviving _ATM_−/− and _ATM_−/− Beclin-1+/− all died of T-cell lymphomas.
Figure 4
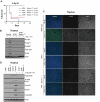
Allelic loss of Beclin-1 does not affect DNA damage responses. (A) Both _ATM_−/− and _ATM_−/− Beclin-1+/− mice display radiosensitivity. Survival of control, _ATM_−/−, and _ATM_−/− Beclin-1+/− mice exposed to 8-Gy total body irradiation is shown (P = .85 between _ATM_−/− and _ATM_−/− Beclin-1+/− mice, log-rank (Mantel-Cox) test. (B-D) The DDR pathway is equally defective in _ATM_−/− and _ATM_−/− Beclin-1+/− mice. (B) Immunoblotting of total protein, phosphorylation status, or both of various ATM substrates, including P-KAP1, P-SMC1, total p53, P-p53, and P-ATM itself in thymocytes from mice sacrificed 2 hours after being exposed to 6-Gy irradiation. Actin was used as loading control. (C) Thymic tissues were isolated from nonirradiated or whole body–irradiated animals (6 Gy/1 hour) and embedded in paraffin followed by immunofluorescence staining against the genomic instability marker γH2AX (green). 4,6-diamidino-2-phenylindole (blue) was used to visualize nuclei (magnification, 40×0.055). (D) The lack of rescue of the DDR abnormalities by Beclin-1 heterozygosity is not because of a delay of DDR signaling. Animals were irradiated with 6 Gy, and tissues were harvested 7 hours later followed by isolation of total proteins from thymocytes. The expression or phosphorylation status of proteins involved in DDR (P-KAP, total-p53, P-p53, and P-ATM) was analyzed. Both _ATM_−/− and _ATM_−/− Beclin-1+/− thymocytes showed a mild increase in the levels of proteins involved in DDR (P-KAP, total-p53, and P-p53) compared with cells exposed for 2 hours (B), but no difference was observed between these 2 genotypes at this time point (D). Actin served as loading control.
Figure 5
Allelic loss of Beclin-1 rescues mitochondrial abnormalities in _ATM_-null thymocytes. (A) Allelic loss of Beclin-1 partially reverses the increase in mitochondrial mass observed in _ATM_−/− mice. Freshly isolated thymic cells were stained with 5nM NAO dye. A representative histogram of NAO fluorescence intensity is illustrated. Graphs illustrate the average mean intensity of NAO for each genotype (n ≥ 3/genotype). (B) Beclin-1 heterozygosity reverses the abnormal increase in mtDNA observed in _ATM_−/− thymocytes. The ratio of mitochondrial/nuclear DNA was quantified in viable thymic cells of mice with desired genotypes by real-time PCR using primers for mitochondrial ND2 and nuclear 18S rRNA genes. Mean values are shown (n ≥ 3/genotype). (C-D) Beclin-1 heterozygosity reverses defects in complex I activity and mROS levels in _ATM_−/− thymocytes. Complex I activity (C) and mROS levels (D) were measured as described in Figure 1. For complex I activity, the relative activities of desired genotypes were graphed. For mROS, a representative histogram of MitoSOX fluorescence intensity per genotype is illustrated. Graph shows the averaged mean intensity for each genotype (n ≥ 3/group). P values were obtained by implementing the Student t test analysis in experiments shown in panels A-E (*P ≤ .04, **P ≤ .01, ***P ≤ .001). (E) Allelic loss of Beclin-1 rescues the increase in cellular respiration observed in the _ATM_−/− mice. The rate of oxygen consumption was measured in viable thymic cells of desired genotypes (n = 3/group). Shown is the OCR of thymic cells as function of time in minutes. Equal numbers of viable cells were used per sample for these experiments. (F-G) Allelic loss of Beclin-1 reduces mitochondrial ultra-structure abnormalities in _ATM_-null thymocytes. (F) Whole thymii of 5- to 8-week-old mice of the indicated genotypes were analyzed by transmission electron microscopy, and representative photomicrographs are shown. Mitochondria with disrupted cristae structure and with swollen appearance were considered abnormal. At least 15 images (magnification, ×10 000) were scored per sample. Red arrowheads illustrate abnormal mitochondria having disorganized cristae structure seen in _ATM_−/− thymic cells. The black arrow denotes an autophagic vacuole. (G) Percentage of abnormal mitochondria quantified in the whole thymus of mice shown in panel F. Each dot, triangle, or square represents a single mouse. Statistical analyses were performed using the Student t test (*P ≤ .03).
Figure 6
ATM localizes to mitochondria and becomes activated by mitochondrial dysfunction without evidence of DNA damage. (A) ATM protein localizes to mitochondria. Normal human fibroblasts were fractionated into nuclear (N), cytoplasmic (C), and mitochondrial (M) fractions and the basal levels of total and phospho-ATM (S1981) were analyzed by immunoblotting. Topoisomerase I (Topo), lactate dehydrogenase (LDH), and COX-IV antibodies were used as N, C, and M markers, respectively. We also analyzed 10 μg of total (T) protein. (B) ATM is activated in response to dysfunctional mitochondria without evidence of DNA damage induction. Viable thymic cells isolated from 6- to 8-week-old wild-type (WT) or _ATM_-null mice were either treated with DMSO or 50μM of the mitochondrial uncoupler CCCP for 3 hours followed by total protein isolation and analysis by immunoblotting for various ATM substrates, including P-KAP1, P-SMC1, total p53, P-p53, and P-ATM itself. As a positive control for ATM activity in vivo, total protein lysates from irradiated (6 Gy) or nonirradiated control mice were included in the assay. LC3-II also was analyzed to confirm increased basal autophagy in _ATM_-null cells and induction of this process by CCCP treatment. (C-D) Transient inhibition of ATM results in mitochondrial mass abnormalities. (C) Wild-type thymocytes were treated with DMSO alone or with 10μM CP (ATM kinase inhibitor) for 3 or 5 hours before staining with MitoTracker Green FM (MTG) for 15 minutes. Flow cytometry histograms of 10 000 viable cells per sample are shown. (D) Control or A-T human fibroblasts were treated with either DMSO or 10μM CP for 3 hours before staining with MTG dye and analysis by flow cytometry. Shown is the mean of fluorescence intensity (MFI) in each sample.
Similar articles
- A new role for ATM: regulating mitochondrial function and mitophagy.
Valentin-Vega YA, Kastan MB. Valentin-Vega YA, et al. Autophagy. 2012 May 1;8(5):840-1. doi: 10.4161/auto.19693. Epub 2012 May 1. Autophagy. 2012. PMID: 22617444 Free PMC article. - Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis.
Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Eaton JS, et al. J Clin Invest. 2007 Sep;117(9):2723-34. doi: 10.1172/JCI31604. J Clin Invest. 2007. PMID: 17786248 Free PMC article. - Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells.
Ambrose M, Goldstine JV, Gatti RA. Ambrose M, et al. Hum Mol Genet. 2007 Sep 15;16(18):2154-64. doi: 10.1093/hmg/ddm166. Epub 2007 Jul 2. Hum Mol Genet. 2007. PMID: 17606465 - ATM and the molecular pathogenesis of ataxia telangiectasia.
McKinnon PJ. McKinnon PJ. Annu Rev Pathol. 2012;7:303-21. doi: 10.1146/annurev-pathol-011811-132509. Epub 2011 Oct 24. Annu Rev Pathol. 2012. PMID: 22035194 Review. - Mitochondria at the crossroads of ATM-mediated stress signaling and regulation of reactive oxygen species.
Lee JH, Paull TT. Lee JH, et al. Redox Biol. 2020 May;32:101511. doi: 10.1016/j.redox.2020.101511. Epub 2020 Mar 21. Redox Biol. 2020. PMID: 32244177 Free PMC article. Review.
Cited by
- Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury.
Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Ma X, et al. Circulation. 2012 Jun 26;125(25):3170-81. doi: 10.1161/CIRCULATIONAHA.111.041814. Epub 2012 May 16. Circulation. 2012. PMID: 22592897 Free PMC article. - Phenformin and ataxia-telangiectasia mutated inhibitors synergistically co-suppress liver cancer cell growth by damaging mitochondria.
Wu T, Zhou S, Qin M, Tang J, Yan X, Huang L, Huang M, Deng J, Xiao D, Hu X, Wu J, Yang X, Li G. Wu T, et al. FEBS Open Bio. 2021 May;11(5):1440-1451. doi: 10.1002/2211-5463.13152. Epub 2021 Apr 3. FEBS Open Bio. 2021. PMID: 33742560 Free PMC article. - Lipopolysaccharide induces placental mitochondrial dysfunction in murine and human systems by reducing MNRR1 levels via a TLR4-independent pathway.
Purandare N, Kunji Y, Xi Y, Romero R, Gomez-Lopez N, Fribley A, Grossman LI, Aras S. Purandare N, et al. iScience. 2022 Oct 12;25(11):105342. doi: 10.1016/j.isci.2022.105342. eCollection 2022 Nov 18. iScience. 2022. PMID: 36339251 Free PMC article. - Metabolic Stress and Mitochondrial Dysfunction in Ataxia-Telangiectasia.
Subramanian GN, Yeo AJ, Gatei MH, Coman DJ, Lavin MF. Subramanian GN, et al. Antioxidants (Basel). 2022 Mar 28;11(4):653. doi: 10.3390/antiox11040653. Antioxidants (Basel). 2022. PMID: 35453338 Free PMC article. Review. - NMR- and MD simulation-based structural characterization of the membrane-associating FATC domain of ataxia telangiectasia mutated.
Abd Rahim MS, Cherniavskyi YK, Tieleman DP, Dames SA. Abd Rahim MS, et al. J Biol Chem. 2019 Apr 26;294(17):7098-7112. doi: 10.1074/jbc.RA119.007653. Epub 2019 Mar 13. J Biol Chem. 2019. PMID: 30867195 Free PMC article.
References
- Lavin MF, Shiloh Y. Ataxia-telangiectasia–a multifaceted genetic disorder associated with defective signal transduction. Curr Opin Immunol. 1996;8:459–464. - PubMed
- Herzog K-H, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. - PubMed
- Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. - PubMed
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9(10):759–769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA076379/CA/NCI NIH HHS/United States
- R01 CA157216/CA/NCI NIH HHS/United States
- CA157216/CA/NCI NIH HHS/United States
- R01 CA076379/CA/NCI NIH HHS/United States
- P30CA21765/CA/NCI NIH HHS/United States
- R01 CA071387/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- CA71387/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous