Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion - PubMed (original) (raw)
Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion
Caroline Bruns et al. J Cell Biol. 2011.
Abstract
The endoplasmic reticulum (ER)-Golgi-independent, unconventional secretion of Acb1 requires many different proteins. They include proteins necessary for the formation of autophagosomes, proteins necessary for the fusion of membranes with the endosomes, proteins of the multivesicular body pathway, and the cell surface target membrane SNARE Sso1, thereby raising the question of what achieves the connection between these diverse proteins and Acb1 secretion. In the present study, we now report that, upon starvation in Saccharomyces cerevisiae, Grh1 is collected into unique membrane structures near Sec13-containing ER exit sites. Phosphatidylinositol 3 phosphate, the ESCRT (endosomal sorting complex required for transport) protein Vps23, and the autophagy-related proteins Atg8 and Atg9 are recruited to these Grh1-containing membranes, which lack components of the Golgi apparatus and the endosomes, and which we call a novel compartment for unconventional protein secretion (CUPS). We describe the cellular proteins required for the biogenesis of CUPS, which we believe is the sorting station for Acb1's release from the cells.
Figures
Figure 1.
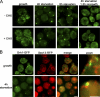
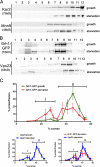
Relocalization of Grh1-GFP upon starvation. (A) Yeast expressing Grh1-GFP grown in normal growth medium were washed and cultured in starvation medium with or without cycloheximide (CHX) for 4 or 8 h. The cells cultured in starvation medium for 4 h were cultured in growth medium for 1.5 h. The cells were visualized by fluorescence microscopy to monitor the localization of Grh1-GFP. (B) Yeast coexpressing Sec13-RFP and Grh1-GFP cultured in growth conditions or starved for 4 h were visualized by fluorescence microscopy. (right) Image at higher magnification. Bars, 2 µm.
Figure 2.
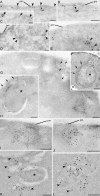
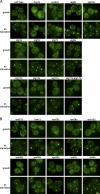
Grh1 localizes near ER exit sites and relocalizes to cup-shaped structures upon starvation. (A–L) Immunoelectron microscopy of ultrathin cryosections of yeast cells. Growth conditions (A–E), 4-h starvation conditions (G–L), and conventional morphology at 4-h starvation (F) are shown. (A–C) Double immunogold labeling of Grh1-GFP (12 nm) and Sec13-RFP (6 nm). (D) Single labeling of Grh1-GFP. (E) Single labeling of Sec13-RFP. (F) Conventional morphology. (G–J) Labeling of Grh1-GFP. (K and L) Double labeling of Grh1-GFP (12 nm) and Sec13-RFP (6 nm). Arrowheads indicate 6-nm gold particles. Arrows indicate membrane lamellae. Asterisks indicate preautophagosomal structure. pm, plasma membrane; v, vacuole. Bars: (A–D, G–I, K, and L) 200 nm; (F) 500 nm; (E and J) 100 nm.
Figure 3.
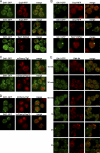
Grh1-GFP does not colocalize with marker proteins of the Golgi apparatus and endosomes under normal growth conditions or upon starvation. (A and B) Grh1-GFP was coexpressed with the early Golgi marker proteins CopI-RFP and AnpI-RFP or the late Golgi marker protein Sec7-DsRed. Cells were cultured in growth conditions (A) or nutrient starved for 4 h (B) and visualized by fluorescence microscopy. (C and D) Grh1-GFP was coexpressed with mCherry-Tlg1 or mCherry-Pep12 in growth conditions (C) or nutrient starved (D) and visualized by fluorescence microscopy. (E) Grh1-GFP–expressing cells were labeled with FM 4-64 and transferred to starvation medium for up to 3 h to visualize the localization of the internalized FM 4-64 with reference to Grh1-GFP. Bar, 2 µm.
Figure 4.
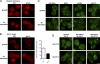
Grh1 colocalizes with Vps23 (ESCRT-I), PI3P, and components of autophagy machinery upon starvation. (A and B) Grh1-GFP was coexpressed with Vps23-mCherry (A) or with Snf7-RFP (B), cultured in growth conditions or nutrient starved for 4 h, and visualized by fluorescence microscopy. The zoom images of A represent high magnification images. (C and D) Grh1-GFP was coexpressed with RFP-Atg9, mCherry-Atg8, and a DsRed-tagged FYVE domain and cultured in growth conditions (C) or nutrient starved (D) and visualized by fluorescence microscopy. The right images represent one of the CUPS at high magnification. (E) The percentages of colocalization were quantified with respect to Grh1 or the indicated marker. At least 60 cells per marker were assessed, and errors are represented as SEM. Bars, 2 µm.
Figure 5.
CUPS is separated from the ER and Golgi membranes. (A and B) Yeast cells cultured in growth conditions or starved for 3 h were fractionated on a continuous 15–60% sucrose gradient for 18 h. The gradient fractions were Western blotted to detect the ER protein Kar2 and the Golgi protein Mnn9 (A) or Vps23 and GFP to monitor Grh1-GFP (B). The numbers indicate the fractions. (C) The percentage of protein contained in each fraction was plotted against the sucrose concentration, and the error bars represent the results and SEM from three independent experiments. The fractions marked ‘I’ represent the starvation-specific pool of Grh1 and Vps23, and the denser fractions ‘II’ represent the ER–Golgi pool of Grh1-GFP and the endosomal pool of Vps23.
Figure 6.
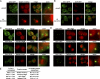
The role of Bug1, autophagy-related proteins, and the ESCRT machinery in CUPS formation. (A and B) Wild-type yeast, yeast strains deleted for BUG1, VPS34, ATG1, 5, 7, 8, 9, 11, 14, 17, 18, and 11/17 (A) or yeast cells deleted for components of the ESCRT machinery (VPS27, HSE1, VPS23, VPS28, MVB12, VPS36, VPS25, VPS20, VPS2, and VPS4; B) expressing Grh1-GFP were cultured in growth medium or starved for 4 h, and the formation of CUPS was visualized by fluorescence microscopy. Bar, 2 µm.
Figure 7.
NSF is not essential for CUPS biogenesis. (A) Wild-type (WT) and grh1Δ yeast expressing Vps23-mCherry were cultured in growth conditions or starved for 4 h and analyzed by fluorescence microscopy. (B) Wild-type and grh1Δ strains expressing RFP-Atg9 were cultured in growth conditions or starved and visualized by fluorescence microscopy. Starved cells showing RFP-Atg9–specific punctate elements representing the CUPS were counted in wild-type and grh1Δ cells. 59 ± 3.3% of wild-type cells showed a localization of Atg9 to CUPS compared with only 28.5 ± 1.8% upon deletion of GRH1 (statistically significant with P < 0.0001; error bars represent SEM). (C) Grh1-GFP expressing wild-type yeast and deleted for TLG2, YPT6, and SSO1 were cultured in growth or starvation medium and visualized by fluorescence microscopy. (D) Grh1-GFP was expressed in sec12-4 and sec18-1 strains. Yeast cells were grown at permissive temperature in growth medium and starved for 3 h at either the permissive or the nonpermissive temperature and visualized by fluorescence microscopy. Bar, 2 µm.
Figure 8.
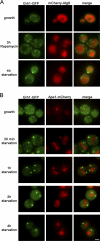
CUPS is distinct from PAS. (A) Yeast cells coexpressing Grh1-GFP and mCherry-Atg8 were cultured in growth medium and either nutrient starved for 4 h or treated with 0.4 µg/ml rapamycin in nutrient-rich medium for 3 h. (B) Yeast cells coexpressing Grh1-GFP and Ape1-mCherry were cultured in growth medium or starved for the indicated time points and visualized by fluorescence microscopy. Bar, 2 µm.
Similar articles
- Unconventional secretion of Acb1 is mediated by autophagosomes.
Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Duran JM, et al. J Cell Biol. 2010 Feb 22;188(4):527-36. doi: 10.1083/jcb.200911154. Epub 2010 Feb 15. J Cell Biol. 2010. PMID: 20156967 Free PMC article. - Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae.
Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Reggiori F, et al. Mol Biol Cell. 2004 May;15(5):2189-204. doi: 10.1091/mbc.e03-07-0479. Epub 2004 Mar 5. Mol Biol Cell. 2004. PMID: 15004240 Free PMC article. - ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion.
Curwin AJ, Brouwers N, Alonso Y Adell M, Teis D, Turacchio G, Parashuraman S, Ronchi P, Malhotra V. Curwin AJ, et al. Elife. 2016 Apr 26;5:e16299. doi: 10.7554/eLife.16299. Elife. 2016. PMID: 27115345 Free PMC article. - Autophagosome biogenesis comes out of the black box.
Chang C, Jensen LE, Hurley JH. Chang C, et al. Nat Cell Biol. 2021 May;23(5):450-456. doi: 10.1038/s41556-021-00669-y. Epub 2021 Apr 26. Nat Cell Biol. 2021. PMID: 33903736 Free PMC article. Review. - Mechanisms of autophagosome biogenesis.
Rubinsztein DC, Shpilka T, Elazar Z. Rubinsztein DC, et al. Curr Biol. 2012 Jan 10;22(1):R29-34. doi: 10.1016/j.cub.2011.11.034. Curr Biol. 2012. PMID: 22240478 Review.
Cited by
- The LC3-conjugation machinery specifies cargo loading and secretion of extracellular vesicles.
Delorme-Axford E, Klionsky DJ. Delorme-Axford E, et al. Autophagy. 2020 Jul;16(7):1169-1171. doi: 10.1080/15548627.2020.1760057. Epub 2020 May 13. Autophagy. 2020. PMID: 32401566 Free PMC article. - Unconventional secretion factor GRASP55 is increased by pharmacological unfolded protein response inducers in neurons.
van Ziel AM, Largo-Barrientos P, Wolzak K, Verhage M, Scheper W. van Ziel AM, et al. Sci Rep. 2019 Feb 7;9(1):1567. doi: 10.1038/s41598-018-38146-6. Sci Rep. 2019. PMID: 30733486 Free PMC article. - Dual roles for autophagy: degradation and secretion of Alzheimer's disease Aβ peptide.
Nilsson P, Saido TC. Nilsson P, et al. Bioessays. 2014 Jun;36(6):570-8. doi: 10.1002/bies.201400002. Epub 2014 Apr 8. Bioessays. 2014. PMID: 24711225 Free PMC article. Review. - MERTK-Mediated LC3-Associated Phagocytosis (LAP) of Apoptotic Substrates in Blood-Separated Tissues: Retina, Testis, Ovarian Follicles.
Yefimova MG, Ravel C, Rolland AD, Bourmeyster N, Jégou B. Yefimova MG, et al. Cells. 2021 Jun 9;10(6):1443. doi: 10.3390/cells10061443. Cells. 2021. PMID: 34207717 Free PMC article. Review. - Spatiotemporal dissection of the Golgi apparatus and the ER-Golgi intermediate compartment in budding yeast.
Tojima T, Suda Y, Jin N, Kurokawa K, Nakano A. Tojima T, et al. Elife. 2024 Mar 19;13:e92900. doi: 10.7554/eLife.92900. Elife. 2024. PMID: 38501165 Free PMC article.
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous