Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2 - PubMed (original) (raw)
Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2
Nyam-Osor Chimge et al. Breast Cancer Res. 2011.
Abstract
Introduction: In contrast to its role in breast cancer (BCa) initiation, estrogen signaling has a protective effect in later stages, where estrogen receptor (ER)α loss associates with aggressive metastatic disease. We asked whether the beneficial effect of estrogen signaling in late-stage BCa is attributable to the recently reported estrogen-mediated antagonism of the pro-metastatic transcription factor Runx2.
Methods: MCF7/Rx2dox breast cancer cells were engineered with a lentivirus expressing Runx2 in response to doxycycline (dox). Cells treated with dox and/or estradiol (E2) were subjected to genome-wide expression profiling, RT-qPCR analysis of specific genes, and Matrigel™ invasion assays. Knockdown of genes of interest was performed using lentiviruses expressing appropriate shRNAs, either constitutively or in response to dox. Gene expression in BCa tumors was investigated using a cohort of 557 patients compiled from publicly available datasets. Association of gene expression with clinical metastasis was assessed by dichotomizing patients into those expressing genes of interest at either high or low levels, and comparing the respective Kaplan-Meier curves of metastasis-free survival.
Results: Runx2 induced epithelial-mesenchymal transition (EMT) evidenced by acquisition of a fibroblastic morphology, decreased expression of E-cadherin, increased expression of vimentin and invasiveness. Runx2 stimulated SNAI2 expression in a WNT- and transforming growth factor (TGF)β-dependent manner, and knockdown of SNAI2 abrogated the pro-metastatic activities of Runx2. E2 antagonized the pro-metastatic activities of Runx2, including SNAI2 upregulation. In primary BCa tumors, Runx2 activity, SNAI2 expression, and metastasis were positively correlated, and SNAI2 expression was negatively correlated with ERα. However, the negative correlation between SNAI2 and ERα in bone-seeking BCa cells was weaker than the respective negative correlation in tumors seeking lung. Furthermore, the absence of ERα in primary tumors was associated with lung- and brain- but not with bone metastasis, and tumor biopsies from bone metastatic sites displayed the unusual combination of high Runx2/SNAI2 and high ERα expression.
Conclusions: E2 antagonizes Runx2-induced EMT and invasiveness of BCa cells, partly through attenuating expression of SNAI2, a Runx2 target required for mediating its pro-metastatic property. That ERα loss promotes non-osseous metastasis by unleashing Runx2/SNAI2 is supported by the negative correlation observed in corresponding tumors. Unknown mechanisms in bone-seeking BCa allow high Runx2/SNAI2 expression despite high ERα level.
Figures
Figure 1
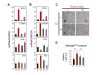
Relationships between Runx2, E2 and SNAI2 in BCa. (A) Runx2 was induced by treating MCF7/Rx2dox BCa cells with dox for 48 hours, and the effect on SNAI2 protein level was assessed by Western blot analysis of nuclear extracts. (B-C) Runx2 was either induced for 48 hours by dox treatment of C4-2B/Rx2dox PCa cells (B) or silenced by 48-hour dox treatment of T47D/shRx2dox BCa cells (C), and the effects on SNAI2 expression were analyzed by RT-qPCR. Western blot analysis of whole cell extracts confirms the respective dox-induced induction and silencing of Runx2. (D) Correlation between SNAI2 mRNA and Runx2 activity in a cohort of 557 BCa tumors. Runx2 activity was defined as the average normalized expression of genes that Runx2 stimulated by ≥ 2 fold in the MCF7/Rx2dox cell culture model, except SNAI2 itself. (E) Expression of SNAI2 mRNA in the same cohort, comparing tumors with high versus low ESR1/ERα levels. Asterisk (*) indicate statistically significant difference (P < 0.05) based on unpaired t-test with Welch correction of the log2-transformation of the signal intensities. (F) Analysis of the tumors with high ESR1/ERα expression for correlation between ESR1/ERα and SNAI2. The patient cohort for D-F was compiled from the publicly available GEO datasets GSE2034, GSE2603 and GSE12276 [37-39]. BCa, breast cancer; dox, doxycycline; E2, estradiol; ESR1/ER, estrogen receptor alpha; GEO, Gene Expression Omnibus; PCa, prostate cancer; RT-qPCR; reverse transcription - quantitative polymerase chain reaction; Runx2, runt related transcription factor 2; SNAI2, snail homolog 2.
Figure 2
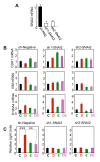
E2 antagonizes Runx2-induced EMT markers and invasiveness of MCF7/Rx2dox BCa cells. (A) MCF7/Rx2dox cells were maintained for two days in medium supplemented with CSS and then treated for two days with either vehicle control (C), dox (D), E2 (E), or both stimulants (DE), followed by RT-qPCR analysis of SNAI2, S100A4, vimentin (VIM) and E-cadherin (CDH1) mRNAs. (B) Relative expression of the same marker genes after treatment as in A for seven days. (C) Phase-contrast images of MCF7/Rx2dox cells after seven days of treatments as indicated. (D) MCF7/Rx2dox cells transduced with lentiviruses constitutively expressing firefly luciferase were placed in BD Biocoat™ Transwell inserts with Matrigel™ and treated for 24 hours as indicated. Luciferase activity in cells that invaded through the Matrigel™ membrane is presented relative to the average control value (Mean ± SD; n = 3; **P < 0.01). CSS, charcoal stripped serum; dox, doxycycline; E2, estradiol; EMT, epithelial-mesenchymal transition; RT-qPCR; reverse transcription - quantitative polymerase chain reaction; Runx2, runt related transcription factor 2; S100A4, S100 calcium binding protein A4; SNAI2, snail homolog 2.
Figure 3
SNAI2 is required for Runx2-mediated EMT. (A) RT-qPCR analysis of SNAI2 mRNA in MCF7/Rx2dox cells transduced with a control (sh-Neg) or each of two independent SNAI2 shRNA lentiviruses. (B-C), MCF7/Rx2dox cells transduced with sh1-SNAI2, sh2-SNAI2, or the negative control virus were treated with vehicle control (C), dox (D), E2 (E), or both stimulants (DE). The effects of SNAI2 knockdown on expression of the indicated EMT markers were assessed by RT-qPCR after seven days and the effects on invasion were assessed using Matrigel™-containing BD Biocoat™ Transwells as in Figure 2D, except dox treatment was initiated 6 hours prior to placing the cells in the transwell chambers. **, P < 0.01; ***, P < 0.005. Dox, doxycycline; EMT, epithelial-mesenchymal transition; E2, estradiol; RT-qPCR, reverse transcription - quantitative polymerase chain reaction; Runx2, runt related transcription factor 2; SNAI2, snail homolog.
Figure 4
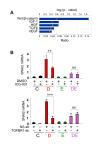
WNT and TGFβ signaling are critical for Runx2-mediated stimulation of SNAI2 expression. (A) The 530 'common' genes, which were regulated by both Runx2 and E2, were analyzed for enrichment for developmental and growth factor signaling pathways using the IPA™ software package. The bars and the upper scale indicate the log2-transformed significance values. The line graph and the bottom scale show magnitudes of enrichment represented as ratios. (B) MCF7/Rx2dox cells were treated for 24 hours with vehicle control (C), dox (D), E2 (E) or both (DE), along with either 10 μM ICG-001, a small molecule inhibitor of Wnt signaling (top), or blocking antibody against the type I TGFβ receptor (bottom). DMSO and a non-specific antibody were used as respective controls. SNAI2 expression was measured by RT-qPCR and corrected for GAPDH. **, P < 0.01; ***, P < 0.005. DMSO, dimethyl sulfoxide; dox, doxycycline; E2, estradiol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HGF, hepatocyte growth factor; IGF, insulin-like growth factor, IPA, Ingenuity Pathway Analysis; RT-qPCR; reverse transcription - quantitative polymerase chain reaction; Runx2, runt related transcription factor 2; TGF, transforming growth factor; SNAI2, snail homolog 2; VEGF, vascular endothelial growth factor.
Figure 5
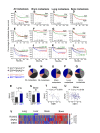
Association of SNAI2 and ERα expression with metastasis. A cohort of 557 BCa tumors compiled from the GEO datasets GSE2034, GSE2603 and GSE12276 [37-39], was dichotomized into groups expressing either high or low levels of either SNAI2 or ESR1/ERα, and Kaplan-Meier curves were constructed for the probability of overall or organ-specific metastasis-free survival in each group. (A-D) Kaplan-Meier curves of overall, brain, lung and bone metastasis-free survival in the cohort of 557 patients according to SNAI2 expression status. (E-H) Kaplan-Meier curves of overall, brain, lung and bone metastasis-free survival in the cohort according to ERα expression status.(I-L) Kaplan-Meier curves of overall, brain, lung and bone metastasis-free survival were generated for the 342 ERα-positive (black and grey curves) and 215 ERα-negative (blue and pink curves) tumors based on high versus low expression of SNAI2. Significance of association was determined by logrank test and the _P_-values are depicted for ERα-positive (green), and ERα-negative tumors (red). (M-Q) Expression status of SNAI2 and ERα in tumors that either have not relapsed (M) or metastasized to any tissue (N), to brain (O), to lung (P), or to bone (Q). (R) Expression of SNAI2 mRNA in lung-seeking tumors expressing ESR1/ERα at high versus low levels. The difference is significant (*, P < 0.05) based on unpaired t-test with Welch correction of the log2-transformed signal intensities. (S) Expression of SNAI2 mRNA in bone-seeking tumors expressing ESR1/ERα at high versus low levels. The difference is insignificant (NS, P = 0.23) based on unpaired t-test with Welch correction of the log2-transformed signal intensities. (T-U) Correlation between SNAI2 mRNA and ESR1/ERα mRNA in lung-seeking (T) and bone-seeking (U) tumors with high ERα. Values represent log2 transformed normalized signal intensities. (V) Standardized intensity plot of RUNX2, SNAI2 and ESR1/ERα gene expressions in 58 human BCa metastases. Expression data of metastatic tissues was extracted from the GEO dataset GSE14020 [9]. BCa, breast cancer; ESR1/ER, estrogen receptor alpha; GEO, Gene Expression Omnibus; RUNX2, runt related transcription factor 2; SNAI2, snail homolog 2.
Similar articles
- Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis.
Ding X, Park SI, McCauley LK, Wang CY. Ding X, et al. J Biol Chem. 2013 Apr 12;288(15):10241-53. doi: 10.1074/jbc.M112.443655. Epub 2013 Feb 27. J Biol Chem. 2013. PMID: 23447531 Free PMC article. - Estrogen receptor-α-miR-1271-SNAI2 feedback loop regulates transforming growth factor-β-induced breast cancer progression.
Liu BW, Yu ZH, Chen AX, Chi JR, Ge J, Yu Y, Cao XC. Liu BW, et al. J Exp Clin Cancer Res. 2019 Mar 1;38(1):109. doi: 10.1186/s13046-019-1112-4. J Exp Clin Cancer Res. 2019. PMID: 30823890 Free PMC article. - The RUNX family in breast cancer: relationships with estrogen signaling.
Chimge NO, Frenkel B. Chimge NO, et al. Oncogene. 2013 Apr 25;32(17):2121-30. doi: 10.1038/onc.2012.328. Epub 2012 Oct 8. Oncogene. 2013. PMID: 23045283 Free PMC article. Review. - Role of RUNX2 in Breast Carcinogenesis.
Wysokinski D, Blasiak J, Pawlowska E. Wysokinski D, et al. Int J Mol Sci. 2015 Sep 2;16(9):20969-93. doi: 10.3390/ijms160920969. Int J Mol Sci. 2015. PMID: 26404249 Free PMC article. Review.
Cited by
- Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression.
Gröger CJ, Grubinger M, Waldhör T, Vierlinger K, Mikulits W. Gröger CJ, et al. PLoS One. 2012;7(12):e51136. doi: 10.1371/journal.pone.0051136. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251436 Free PMC article. - Runx2 contributes to the regenerative potential of the mammary epithelium.
Ferrari N, Riggio AI, Mason S, McDonald L, King A, Higgins T, Rosewell I, Neil JC, Smalley MJ, Sansom OJ, Morris J, Cameron ER, Blyth K. Ferrari N, et al. Sci Rep. 2015 Oct 22;5:15658. doi: 10.1038/srep15658. Sci Rep. 2015. PMID: 26489514 Free PMC article. - RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer.
Chimge NO, Little GH, Baniwal SK, Adisetiyo H, Xie Y, Zhang T, O'Laughlin A, Liu ZY, Ulrich P, Martin A, Mhawech-Fauceglia P, Ellis MJ, Tripathy D, Groshen S, Liang C, Li Z, Schones DE, Frenkel B. Chimge NO, et al. Nat Commun. 2016 Feb 26;7:10751. doi: 10.1038/ncomms10751. Nat Commun. 2016. PMID: 26916619 Free PMC article. - Detection of RUNX2 gene expression in cumulus cells in women undergoing controlled ovarian stimulation.
Papamentzelopoulou M, Mavrogianni D, Dinopoulou V, Theofanakis H, Malamas F, Marinopoulos S, Bletsa R, Anagnostou E, Kallianidis K, Loutradis D. Papamentzelopoulou M, et al. Reprod Biol Endocrinol. 2012 Nov 28;10:99. doi: 10.1186/1477-7827-10-99. Reprod Biol Endocrinol. 2012. PMID: 23186169 Free PMC article. - Clinical significance of RUNX2 expression in patients with nonsmall cell lung cancer: a 5-year follow-up study.
Li H, Zhou RJ, Zhang GQ, Xu JP. Li H, et al. Tumour Biol. 2013 Jun;34(3):1807-12. doi: 10.1007/s13277-013-0720-4. Epub 2013 Mar 8. Tumour Biol. 2013. PMID: 23471668
References
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials