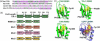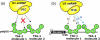Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1 - PubMed (original) (raw)
Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1
William J Bauer et al. J Mol Biol. 2012.
Abstract
T-cell intracellular antigen-1 (TIA-1) regulates developmental and stress-responsive pathways through distinct activities at the levels of alternative pre-mRNA splicing and mRNA translation. The TIA-1 polypeptide contains three RNA recognition motifs (RRMs). The central RRM2 and C-terminal RRM3 associate with cellular mRNAs. The N-terminal RRM1 enhances interactions of a C-terminal Q-rich domain of TIA-1 with the U1-C splicing factor, despite linear separation of the domains in the TIA-1 sequence. Given the expanded functional repertoire of the RRM family, it was unknown whether TIA-1 RRM1 contributes to RNA binding as well as documented protein interactions. To address this question, we used isothermal titration calorimetry and small-angle X-ray scattering to dissect the roles of the TIA-1 RRMs in RNA recognition. Notably, the fas RNA exhibited two binding sites with indistinguishable affinities for TIA-1. Analyses of TIA-1 variants established that RRM1 was dispensable for binding AU-rich fas sites, yet all three RRMs were required to bind a polyU RNA with high affinity. Small-angle X-ray scattering analyses demonstrated a "V" shape for a TIA-1 construct comprising the three RRMs and revealed that its dimensions became more compact in the RNA-bound state. The sequence-selective involvement of TIA-1 RRM1 in RNA recognition suggests a possible role for RNA sequences in regulating the distinct functions of TIA-1. Further implications for U1-C recruitment by the adjacent TIA-1 binding sites of the fas pre-mRNA and the bent TIA-1 shape, which organizes the N- and C-termini on the same side of the protein, are discussed.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Fig. 1
(a) Nucleotide sequence comparisons of RNA sites regulated by TIA-1, either adjacent the 5′ splice sites (SS) of fas intron 5, fas intron 6, or msl-2 intron 1, or an AU-rich region within the tnf-α 3′ UTR. Adenosines are colored green, and uridines are magenta. Oligonucleotides used in this study (underlined and in bold) were derived from the 5′ splice site of fas intron 5 for calorimetry or the minimal AU-element of _tnf_-α RNA for SAXS. (b) Schematic diagram of full length TIA-1 and the RRM123, Mut1, Mut2, and Mut3 constructs used in this study. (c) TIA-1 or TIAR RRM structures compared with the second U2AF65 RRM bound to a polyU-tract (TIA-1 RRM2 PDB ID 3BS9; TIAR RRM1 PDB ID 2CQI, TIAR PDB ID RRM3 1×4G; U2AF65 RRM2 PDB ID 2G4B). For clarity, the TIAR residues are numbered according to human TIA-1 sequence. The RNP1 and RNP2 consensus residues that were altered by site-directed mutagenesis are labeled in (b) and (c).
Fig. 2
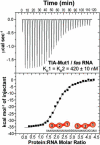
Comparison of wild-type TIA-1 (wtTIA-RRM123) binding RNA sites. Representative isotherms for (a) wtTIA-RRM123 (140 μM) titrated into fas RNA (7 μM) and fit with an identical binding sites model. A nonidentical sites model produced similar quality fit. (b) wtTIA-RRM123 (70 μM) into U20 RNA (7 μM). The curve resulting from the model of nonidentical sites is shown in black, whereas that of identical sites is overlaid in red. The KD values and proposed models for each binding event are inset. The RRM domains of each TIA-1 molecule are numbered from N- to C-terminus here and throughout. The enthalpy and entropy changes of binding are given in Table 1. (c–d) Qualitative electrophoretic mobility shift assays demonstrate the formation of two complexes during titration of wtTIA-RRM123 into fluorescein-labeled (c) fas or (d) U20 RNA sites (2.5 μM). The final protein concentrations were 0.7, 1.1, 1.8, 2.9, 4.6, 11.5 and 18.5 μM for the fas RNA titration, and 0.6, 1.0, 1.6, 2.6, 6.1, 9.7 and 15.6 μM for the U20 RNA titration.
Fig. 3
Comparison of U20 RNA binding to TIA-1 variants. Representative isotherms for the titration of (a) U20 RNA (70 μM) into wtTIA-RRM123 (7 μM), (b) U20 RNA (42 μM) into TIA-Mut1 (6 μM), (c) U20 RNA (80 μM) into TIA-Mut2 (9 μM), (d) U20 RNA (50 μM) into TIA-Mut3 (5 μM). TIA-Mut1 was fit with an identical sites model, since the fit of a nonidentical sites model was similar. The wtTIARRM123, TIA-Mut2 and TIA-Mut3 were fit with a nonidentical sites model (black line). For comparison, the fits of the identical sites models are overlaid (red line). The KD values and proposed models for the saturated RNA complexes are inset. The enthalpy and entropy changes of the titrations are given in Table 1.
Fig. 4
Representative isotherm for TIA-Mut1 (140 μM) titrated into fas RNA (7 μM) fit with an identical binding sites model.
Fig. 5
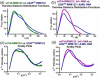
Small-angle X-ray scattering data and ab initio models for wtTIA-RRM123 in the absence (green) and presence (purple) of _tnf_-α RNA site (5′-UUAUUUAUUUA-3′). (a) The experimental X-ray scattering profiles displaced by arbitrary units along the y-axis for clarity. The fits of theoretical scattering from the most typical ab initio models generated using DAMMIN are overlaid (dashed lines). (b) The P(r) functions were scaled to equalize the areas under the integrated functions. (c) Kratky plots were calculated after scaling the scattering intensities. The averaged and filtered DAMMIN envelopes of (d) wtTIA-RRM123 and (e) wtTIA-RRM123 / _tnf_-α RNA were overlaid with the most typical BUNCH rigid body models of the individual RRMs (PDB codes given in Fig. 1c), and the envelopes were superimposed in (f). The radii of gyration (Rg), maximum intraparticle sizes (Dmax), χ2 and NSD values are given in Table 2. Scattering data were collected at ALS.
Fig. 6
Small-angle X-ray scattering data for wtTIA-RRM123 in the absence (green) or presence (purple) of the _tnf_-α RNA site respectively compared with U2AF65 RRM123 in the absence or presence of its RNA site (AdMl Py tract, 5′-UCCCUUUUUUUUCC-3′). (a, b) Pairwise distance distribution functions and (c, d) Kratky plots.
Fig. 7
Small-angle X-ray scattering data for wtTIA-RRM123 (green) compared TIA-Mut1 (red). (a) X-ray scattering plots, (b) pairwise distance distribution functions, and (c) Kratky plots are shown. The averaged and filtered DAMMIN envelopes of (d) wtTIA-RRM123 and (e) TIA-Mut1 are overlaid with the most typical BUNCH rigid body models of the individual RRMs (PDB codes given for Fig. 1c), or (f) superimposed. The radii of gyration (Rg), maximum intraparticle sizes (Dmax), χ2 g and NSD values are given in Table 2. Scattering data were collected at CHESS.
Fig. 8
Models for U1-C interactions with TIA-1. The TIA-1 RRM1 contributes to TIA-1 binding to long uridine tracts (a), but is dispensable for binding tandem AU-tracts such as those following fas exon 5 (b). The latter case may free RRM1 to interact with other molecules such as U1-C. The `V' shape of TIA-1 aligns the N- and C-terminal domains along one side of the molecule, consistent with the intramolecular participation of both domains in U1-C interactions as schematically diagrammed with the (i) brown arrows. The association of two TIA-1 molecules with the AU-rich sequences following fas exon 5 leads to an alternative model for U1-C recruitment to the fas 5′ splice site via intermolecular interactions with TIA-1, as indicated by (ii) blue arrows.
Similar articles
- Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1.
Wang I, Hennig J, Jagtap PK, Sonntag M, Valcárcel J, Sattler M. Wang I, et al. Nucleic Acids Res. 2014 May;42(9):5949-66. doi: 10.1093/nar/gku193. Epub 2014 Mar 25. Nucleic Acids Res. 2014. PMID: 24682828 Free PMC article. - The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain.
Cruz-Gallardo I, Aroca Á, Gunzburg MJ, Sivakumaran A, Yoon JH, Angulo J, Persson C, Gorospe M, Karlsson BG, Wilce JA, Díaz-Moreno I. Cruz-Gallardo I, et al. RNA Biol. 2014;11(6):766-76. doi: 10.4161/rna.28801. Epub 2014 Apr 24. RNA Biol. 2014. PMID: 24824036 Free PMC article. - A structural insight into the C-terminal RNA recognition motifs of T-cell intracellular antigen-1 protein.
Aroca A, Díaz-Quintana A, Díaz-Moreno I. Aroca A, et al. FEBS Lett. 2011 Oct 3;585(19):2958-64. doi: 10.1016/j.febslet.2011.07.037. Epub 2011 Aug 11. FEBS Lett. 2011. PMID: 21846467 - Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1.
Förch P, Valcárcel J. Förch P, et al. Apoptosis. 2001 Dec;6(6):463-8. doi: 10.1023/a:1012441824719. Apoptosis. 2001. PMID: 11595836 Review. - RNA recognition motifs involved in nuclear import of RNA-binding proteins.
Cassola A, Noé G, Frasch AC. Cassola A, et al. RNA Biol. 2010 May-Jun;7(3):339-44. doi: 10.4161/rna.7.3.12087. Epub 2010 May 14. RNA Biol. 2010. PMID: 20458169 Review.
Cited by
- GraphProt: modeling binding preferences of RNA-binding proteins.
Maticzka D, Lange SJ, Costa F, Backofen R. Maticzka D, et al. Genome Biol. 2014 Jan 22;15(1):R17. doi: 10.1186/gb-2014-15-1-r17. Genome Biol. 2014. PMID: 24451197 Free PMC article. - Restriction of an intron size en route to endothermy.
Královičová J, Borovská I, Pengelly R, Lee E, Abaffy P, Šindelka R, Grutzner F, Vořechovský I. Královičová J, et al. Nucleic Acids Res. 2021 Mar 18;49(5):2460-2487. doi: 10.1093/nar/gkab046. Nucleic Acids Res. 2021. PMID: 33550394 Free PMC article. - microRNAs Promoting Growth of Gastric Cancer Xenografts and Correlation to Clinical Prognosis.
Weidle UH, Birzele F, Nopora A. Weidle UH, et al. Cancer Genomics Proteomics. 2021 Jan-Feb;18(1):1-15. doi: 10.21873/cgp.20237. Epub 2021 Jan 8. Cancer Genomics Proteomics. 2021. PMID: 33419892 Free PMC article. Review. - Linking hnRNP Function to ALS and FTD Pathology.
Purice MD, Taylor JP. Purice MD, et al. Front Neurosci. 2018 May 15;12:326. doi: 10.3389/fnins.2018.00326. eCollection 2018. Front Neurosci. 2018. PMID: 29867335 Free PMC article. Review. - Topoisomerase 3β is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction.
Ahmad M, Shen W, Li W, Xue Y, Zou S, Xu D, Wang W. Ahmad M, et al. Nucleic Acids Res. 2017 Mar 17;45(5):2704-2713. doi: 10.1093/nar/gkw1293. Nucleic Acids Res. 2017. PMID: 28039324 Free PMC article.
References
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. - PubMed
- Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J Biol Chem. 2004;279:16980–16988. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous