Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes - PubMed (original) (raw)
. 2011 Dec 11;44(1):53-7.
doi: 10.1038/ng.1031.
Dong Shen, Li Ding, Theresa Okeyo-Owuor, Cara L Lunn, Jin Shao, Kilannin Krysiak, Christopher C Harris, Daniel C Koboldt, David E Larson, Michael D McLellan, David J Dooling, Rachel M Abbott, Robert S Fulton, Heather Schmidt, Joelle Kalicki-Veizer, Michelle O'Laughlin, Marcus Grillot, Jack Baty, Sharon Heath, John L Frater, Talat Nasim, Daniel C Link, Michael H Tomasson, Peter Westervelt, John F DiPersio, Elaine R Mardis, Timothy J Ley, Richard K Wilson, Matthew J Walter
Affiliations
- PMID: 22158538
- PMCID: PMC3247063
- DOI: 10.1038/ng.1031
Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes
Timothy A Graubert et al. Nat Genet. 2011.
Abstract
Myelodysplastic syndromes (MDS) are hematopoietic stem cell disorders that often progress to chemotherapy-resistant secondary acute myeloid leukemia (sAML). We used whole-genome sequencing to perform an unbiased comprehensive screen to discover the somatic mutations in a sample from an individual with sAML and genotyped the loci containing these mutations in the matched MDS sample. Here we show that a missense mutation affecting the serine at codon 34 (Ser34) in U2AF1 was recurrently present in 13 out of 150 (8.7%) subjects with de novo MDS, and we found suggestive evidence of an increased risk of progression to sAML associated with this mutation. U2AF1 is a U2 auxiliary factor protein that recognizes the AG splice acceptor dinucleotide at the 3' end of introns, and the alterations in U2AF1 are located in highly conserved zinc fingers of this protein. Mutant U2AF1 promotes enhanced splicing and exon skipping in reporter assays in vitro. This previously unidentified, recurrent mutation in U2AF1 implicates altered pre-mRNA splicing as a potential mechanism for MDS pathogenesis.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors have no competing interest to declare.
Figures
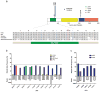
Figure 1. U2AF1 mutations found in patients with myelodysplastic syndromes (MDS)
(a) Missense mutations were detected in codons 34 and 157 of U2AF1. The ZnF1 (zinc finger 1), UHM (U2AF homology motif), ZnF2 (zinc finger 2), and RS (arginine-serine rich) domains are shown. The amino acid sequence of the ZnF1 domain is highly conserved (shaded). The zinc coordinating and mutated residue are shown in blue (asterisks) and red (arrow), respectively. (b) Deep sequencing of U2AF1 using DNA collected from paired normal, MDS, or secondary AML (sAML) samples. Mutant allele frequencies represent the proportion of sequencing reads supporting the mutant allele reads/total reads. Total read counts are shown below (mean 5,651 reads/sample). The mutation is present in the majority of cells (mutant allele frequency 31.4–48.2%) in all cases. (c) Deep sequencing of cDNA from MDS or sAML samples. The mutant allele is expressed in all cases tested. UPN, unique patient number.
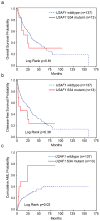
Figure 2. Impact of U2AF1 mutations on clinical outcome
(a) Overall and (b) Event-free survival are not impacted by U2AF1 genotype. (c) The probability of secondary AML (sAML) progression is increased in patients with U2AF1 mutations (P=0.03).
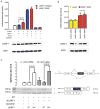
Figure 3. U2AF1 S34F mutation induces splicing alterations
(a) Transient coexpression of the pTN24 double-reporter splicing construct with or without a splicing enhancer (Tra2α), splicing inhibitor (hnRNPG), wild-type U2AF1, or mutant (S34F) U2AF1 cDNA in 293T cells. The double-reporter construct constitutively expresses β-galactosidase, while luciferase is expressed only if an upstream intron which contains multiple stop codons is spliced out. In the absence of the Tra2α splicing enhancer or hnRNPG splicing inhibitor, expression of the mutant U2AF1 increases splicing of the pTN24 construct resulting in an increase in the luciferase/β-galactosidase expression compared to expression of wild-type U2AF1 (P<0.001). Tra2α (positive control) and hnRNPG (negative control) cause increased or decreased splice efficiency, respectively. A representative Western blot of U2AF1 levels in the same cells used for luciferase assays is shown below each combination of plasmids. (b) Transient coexpression of the pTN24 double-reporter splicing construct with a control plasmid (vector) or mutant (S34F) U2AF1 cDNA in the presence of a control siRNA or siRNA targeting the endogenous U2AF1 in 293T cells. Expression of the S34F mutant U2AF1 results in an increase in splicing of the pTN24 construct with an increase in the luciferase/β-galactosidase expression compared to a control plasmid (P<0.001). The result is independent of endogenous U2AF1 expression levels. A representative Western blot of U2AF1 is shown below each condition. (c) The GH1 minigene was transiently transfected into 293T cells with a control plasmid (pcDNA3.1-YFP), wild-type U2AF1 cDNA_,_ or mutant (S34F) U2AF1 cDNA in the presence of a control siRNA or siRNA targeting the endogenous U2AF1. RNA was harvested 48 hours later and a reverse transcriptase (RT) reaction was performed to create cDNA. PCR using the indicated primers resulted in a fully spliced 505 base pair amplicon or a 386 base pair amplicon that skips the middle exon shaded in black (exon skipping). A representative PCR gel image is shown and the ratio of the lower band (exon skipping = amplicon b) relative to the fully spliced upper band (amplicon a) is shown above each condition. Expression of the S34F mutant U2AF1 results in an increase in exon skipping compared to control or wild-type U2AF1 (P<0.02). bp, base pair; WT, wild-type; Mut, mutant; T7, T7 primer. Error bars, s.d.
Similar articles
- U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing.
Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, Kielkopf CL, Ley TJ, Walter MJ, Graubert TA. Okeyo-Owuor T, et al. Leukemia. 2015 Apr;29(4):909-17. doi: 10.1038/leu.2014.303. Epub 2014 Oct 14. Leukemia. 2015. PMID: 25311244 Free PMC article. - U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome.
Qian J, Yao DM, Lin J, Qian W, Wang CZ, Chai HY, Yang J, Li Y, Deng ZQ, Ma JC, Chen XX. Qian J, et al. PLoS One. 2012;7(9):e45760. doi: 10.1371/journal.pone.0045760. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23029227 Free PMC article. - Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors.
Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Je EM, et al. Int J Cancer. 2013 Jul;133(1):260-5. doi: 10.1002/ijc.28011. Epub 2013 Feb 5. Int J Cancer. 2013. PMID: 23280334 - Prognostic value of U2AF1 mutant in patients with de novo myelodysplastic syndromes: a meta-analysis.
Wang H, Zhang N, Wu X, Zheng X, Ling Y, Gong Y. Wang H, et al. Ann Hematol. 2019 Dec;98(12):2629-2639. doi: 10.1007/s00277-019-03843-3. Epub 2019 Nov 21. Ann Hematol. 2019. PMID: 31754743 - Splicing Factor Mutations in Myelodysplasias: Insights from Spliceosome Structures.
Jenkins JL, Kielkopf CL. Jenkins JL, et al. Trends Genet. 2017 May;33(5):336-348. doi: 10.1016/j.tig.2017.03.001. Epub 2017 Mar 31. Trends Genet. 2017. PMID: 28372848 Free PMC article. Review.
Cited by
- HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness.
Ho JS, Di Tullio F, Schwarz M, Low D, Incarnato D, Gay F, Tabaglio T, Zhang J, Wollmann H, Chen L, An O, Chan THM, Hall Hickman A, Zheng S, Roudko V, Chen S, Karz A, Ahmed M, He HH, Greenbaum BD, Oliviero S, Serresi M, Gargiulo G, Mann KM, Hernando E, Mulholland D, Marazzi I, Wee DKB, Guccione E. Ho JS, et al. Elife. 2021 Jun 2;10:e59654. doi: 10.7554/eLife.59654. Elife. 2021. PMID: 34075878 Free PMC article. - A CRISPR RNA-binding protein screen reveals regulators of RUNX1 isoform generation.
Davis AG, Einstein JM, Zheng D, Jayne ND, Fu XD, Tian B, Yeo GW, Zhang DE. Davis AG, et al. Blood Adv. 2021 Mar 9;5(5):1310-1323. doi: 10.1182/bloodadvances.2020002090. Blood Adv. 2021. PMID: 33656539 Free PMC article. - Knockdown of USP39 induces cell cycle arrest and apoptosis in melanoma.
Zhao Y, Zhang B, Lei Y, Sun J, Zhang Y, Yang S, Zhang X. Zhao Y, et al. Tumour Biol. 2016 Oct;37(10):13167-13176. doi: 10.1007/s13277-016-5212-x. Epub 2016 Jul 25. Tumour Biol. 2016. PMID: 27456357 - Aberrant pre-mRNA processing in cancer.
Biswas J, Boussi L, Stein E, Abdel-Wahab O. Biswas J, et al. J Exp Med. 2024 Nov 4;221(11):e20230891. doi: 10.1084/jem.20230891. Epub 2024 Sep 24. J Exp Med. 2024. PMID: 39316554 Free PMC article. Review. - Significance of myelodysplastic syndrome-associated somatic variants in the evaluation of patients with pancytopenia and idiopathic cytopenias of undetermined significance.
Fernandez-Pol S, Ma L, Ohgami RS, Arber DA. Fernandez-Pol S, et al. Mod Pathol. 2016 Sep;29(9):996-1003. doi: 10.1038/modpathol.2016.100. Epub 2016 Jun 3. Mod Pathol. 2016. PMID: 27255165
References
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01CA101937/CA/NCI NIH HHS/United States
- R01 HL082973/HL/NHLBI NIH HHS/United States
- RC2 HL102927/HL/NHLBI NIH HHS/United States
- R01HL082973/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- P01 CA101937/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- RC2HL102927/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous