Identification of components of the host type IA phosphoinositide 3-kinase pathway that promote internalization of Listeria monocytogenes - PubMed (original) (raw)
Identification of components of the host type IA phosphoinositide 3-kinase pathway that promote internalization of Listeria monocytogenes
Shahanawaz Jiwani et al. Infect Immun. 2012 Mar.
Abstract
The bacterial pathogen Listeria monocytogenes causes food-borne illnesses resulting in gastroenteritis, meningitis, or abortion. Listeria promotes its internalization into some human cells through binding of the bacterial surface protein InlB to the host receptor tyrosine kinase Met. The interaction of InlB with the Met receptor stimulates host signaling pathways that promote cell surface changes driving bacterial uptake. One human signaling protein that plays a critical role in Listeria entry is type IA phosphoinositide 3-kinase (PI 3-kinase). The molecular mechanism by which PI 3-kinase promotes bacterial internalization is not understood. Here we perform an RNA interference (RNAi)-based screen to identify components of the type IA PI 3-kinase pathway that control the entry of Listeria into the human cell line HeLa. The 64 genes targeted encode known upstream regulators or downstream effectors of type IA PI 3-kinase. The results of this screen indicate that at least 9 members of the PI 3-kinase pathway play important roles in Listeria uptake. These 9 human proteins include a Rab5 GTPase, several regulators of Arf or Rac1 GTPases, and the serine/threonine kinases phosphoinositide-dependent kinase 1 (PDK1), mammalian target of rapamycin (mTor), and protein kinase C-ζ. These findings represent a key first step toward understanding the mechanism by which type IA PI 3-kinase controls bacterial internalization.
Figures
Fig 1
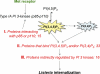
Human type IA PI 3-kinase pathway components targeted in the RNAi-based screen. During infection with Listeria, host type IA PI 3-kinase is activated downstream of the Met receptor and plays a critical role in bacterial internalization (45, 46, 78). Type IA PI 3-kinase uses PI(4,5)P2 as a substrate and produces the lipid second-messenger product PI(3,4,5)P3 (11, 31). PI(3,4)P2 is another second messenger, which is generated from PI(3,4,5)P3 by phosphatases (11). The RNAi-based screen performed in this study targeted three categories of host genes encoding proteins that act on the type IA PI 3-kinase signaling pathway. Category I genes encode proteins that interact with the 85-kDa regulatory and/or 110-kDa catalytic subunit of PI 3-kinase. Category II genes code for proteins that bind to the PI 3-kinase lipid products PI(3,4,5)P3 and/or PI(3,4)P2. Category III genes encode products that are indirectly controlled by type IA PI 3-kinase.
Fig 2
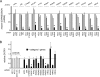
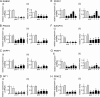
Effects of siRNAs targeting category I host genes on gene expression and InlB-mediated Listeria entry. (A) Inhibition of host gene expression by siRNA pools. HeLa cells were transfected with siRNA pools targeting the indicated category I host genes. As controls, cells were transfected with a nontargeting control siRNA (NTC1) or were mock transfected in the absence of siRNAs (none). Approximately 48 h after transfection, cell lysates were prepared. Gene expression was analyzed by real-time PCR. Relative gene expression values were determined as described in Materials and Methods. Data are means ± SEM for 3 to 5 experiments, depending on the siRNA condition. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1 or no-siRNA control (by the Tukey-Kramer posttest). (B) Impact of siRNA pools on Listeria internalization into host cells. HeLa cells were transfected with siRNA pools directed against the indicated category I host gene. As controls, cells either were transfected with one of two nontargeting control siRNAs (NTC1, NTC2) or with a siRNA directed against lamin A/C or were mock transfected in the absence of siRNAs (none). About 48 h posttransfection, bacterial entry was assessed by using gentamicin protection assays. Relative entry values were obtained by normalization to entry in cells treated with the NTC1 control, as described in Materials and Methods. Data are means ± SEM. Results for siRNAs targeting category I genes are from 3 to 7 experiments, depending on the siRNA condition. Data obtained under the no-siRNA (none), NTC2, or lamin A/C siRNA control condition are from 46, 13, or 13 experiments, respectively. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1, NTC2, lamin A/C, or no-siRNA control (by the Tukey-Kramer posttest).
Fig 3
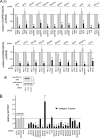
Effects of siRNAs directed against category II host genes on human gene expression and internalization of Listeria. (A) Inhibition of host gene expression. HeLa cells were transfected with siRNA pools targeting the indicated category II host genes. Control transfection conditions and analysis of gene expression were carried out as described in the legend for Fig. 2. (i) Gene expression was analyzed by real-time PCR for all but one of the genes. (ii) In the case of PLCG1 (encoding PLC-γ1 protein), gene expression was assessed by Western blotting, since the probe did not detect expression (see Materials and Methods). Data are means ± SEM for 3 to 5 experiments, depending on the siRNA condition. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1 or no-siRNA control (by the Tukey-Kramer posttest). (B) Effects of siRNA pools on Listeria internalization. HeLa cells were transfected with siRNA pools directed against the indicated category II host gene. Control transfection conditions and measurement of bacterial entry were carried out as described in the legend for Fig. 2. Data for siRNAs targeting category II genes are means ± SEM for 3 to 5 experiments. Data obtained under the no-siRNA (none), NTC2, or lamin A/C siRNA control condition are from 46, 13, or 13 experiments, respectively. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1, NTC2, lamin A/C, or no-siRNA control (by the Tukey-Kramer posttest).
Fig 4
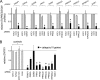
Effects of siRNAs targeting category III host genes on human gene expression and entry of Listeria. (A) Inhibition of host gene expression. HeLa cells were transfected with siRNA pools targeting the indicated category III host genes. Control transfection conditions were those described in the legend for Fig. 2. For experiments with siRNAs targeting category III genes, data are means ± SEM for 3 to 5 experiments, depending on the siRNA condition. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1 or no-siRNA control (by the Tukey-Kramer posttest). (B) Impact of siRNA pools on Listeria entry. HeLa cells were transfected with siRNA pools directed against the indicated category III host genes. Control transfection conditions and measurement of bacterial entry were carried out as described in the legend for Fig. 2. Data for siRNAs targeting category III genes are means ± SEM for 3 to 7 experiments. Data obtained under the no-siRNA (none), NTC2, or lamin A/C siRNA control condition are from 46, 13, or 13 experiments, respectively. Statistical analysis by ANOVA gave a P value of <0.0001. *, P < 0.05 relative to the NTC1, NTC2, lamin A/C, or no-siRNA control (by the Tukey-Kramer posttest).
Fig 5
Multiple siRNAs inhibiting target gene expression impair Listeria internalization. HeLa cells were transfected with siRNA pools or with an individual siRNA targeting the human gene indicated in each panel. Four individual siRNAs were tested for PDPK1 and FRAP1, whereas three siRNAs were tested for all other genes. As controls, cells either were mock transfected in the absence of siRNA (no siRNA) (open bars) or were transfected with control nontargeting siRNA 1 (NTC1) (shaded bars). (i) Effects of siRNAs on host gene expression. Gene expression was analyzed by real-time PCR. Data are means ± SEM for 3 to 4 experiments, depending on the siRNA condition. ANOVA gave P values of <0.0001 for RAB5C (A), PDPK1 (E), SWAP70 (F), FRAP1 (G), and PRKCZ (H), 0.0001 for GIT1 (D), 0.0003 for DAPP1 (C), and 0.0012 for PSCD2 (B). *, P < 0.05 relative to the no-siRNA or NTC1 control (by the Tukey-Kramer posttest). (ii) Effects of siRNA pools on Listeria internalization. Data are means ± SEM for 3 to 7 experiments, depending on the siRNA condition. Statistical analysis by ANOVA gave P values of <0.0001 for all data in all panels. *, P < 0.05 relative to the no-siRNA or NTC1 control (by the Tukey-Kramer posttest).
Fig 6
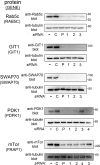
Confirmation that siRNAs inhibit expression at the protein level. The human proteins evaluated for expression are given on the left, followed by the gene names in parentheses. HeLa cells were transfected with siRNA pools (P) or with individual siRNAs directed against the indicated human gene. As controls, cells either were mock transfected in the absence of siRNA (−) or were transfected with nontargeting control 1 siRNA (C). Approximately 48 h after transfection, cells were solubilized in RIPA buffer. The indicated target protein was detected by Western blotting as described in Materials and Methods. In order to confirm equivalent loading, membranes were then stripped and were probed a second time, with anti-tubulin antibodies.
Fig 7
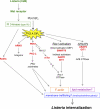
Potential mechanisms of control of Listeria entry by the type IA PI 3-kinase signaling pathway. Infection of human cells with Listeria expressing InlB results in activation of the host Met receptor and of type IA PI 3-kinase (24, 45, 46, 78). The RNAi-based screen described in this work led to the identification of nine human proteins involved in PI 3-kinase signaling that play important roles in Listeria entry. Based on the biological functions of these nine proteins reported in the scientific literature, a diagram was constructed depicting some of the possible ways in which the host proteins could participate in bacterial uptake. Rab5c, a protein that interacts with regulatory and catalytic subunits of type IA PI 3-kinase, could promote Listeria entry by controlling the host endocytic machinery (14, 85). ARNO, an activator of Arf GTPases that binds directly to the PI 3-kinase product PI(3,4,5)P3, might help maintain proper levels of integrins, a class of receptor recently found to enhance InlB-mediated entry, in the plasma membrane (2). The serine/threonine kinase PKC-ζ could promote Listeria internalization by controlling the actin cytoskeleton and/or the delivery of membrane through exocytosis (7, 53, 73). PKC-ζ is indirectly regulated by PI 3-kinase through the master kinase PDK1, which is a direct target of PI(3,4,5)P3 (3). mTor, a serine/threonine kinase indirectly controlled by type IA PI 3-kinase, might promote bacterial uptake through activation of the host proteins Rac1 and/or PKC-α (not shown) (75, 91). Apart from mTor, three other human proteins identified in the RNAi screen have the potential to control Listeria entry though activation of Rac1 GTPase. These proteins, SWAP70, DAPP1, and GIT1, each bind directly to PI(3,4,5)P3. SWAP70 is a direct activator of Rac1 and stimulates nucleotide exchange on the GTPase (79). DAPP1 and GIT1 lack recognizable guanine nucleotide exchange factor (GEF) domains and likely activate Rac1 indirectly. In addition to being an indirect activator of Rac1, GIT1 is also a GTPase-activating protein (GAP) that inhibits Arf6 GTPase (41). Along with ARAP2, another Arf6 GAP needed for Listeria entry (34), GIT1 might restrain the activation of Arf6, which would otherwise interfere with bacterial uptake. Constitutively activated Arf6 alleles inhibit Listeria internalization (34) and also induce the redistribution of cholesterol from the plasma membrane to internal membrane compartments (65). Since plasma membrane cholesterol is critical for InlB-mediated entry, it is possible that GIT1 and/or ARAP2 promotes Listeria uptake by maintaining proper localization of cholesterol and/or other lipids (34).
Similar articles
- Host Serine/Threonine Kinases mTOR and Protein Kinase C-α Promote InlB-Mediated Entry of Listeria monocytogenes.
Bhalla M, Law D, Dowd GC, Ireton K. Bhalla M, et al. Infect Immun. 2017 Jun 20;85(7):e00087-17. doi: 10.1128/IAI.00087-17. Print 2017 Jul. Infect Immun. 2017. PMID: 28461391 Free PMC article. - Critical role for the host GTPase-activating protein ARAP2 in InlB-mediated entry of Listeria monocytogenes.
Gavicherla B, Ritchey L, Gianfelice A, Kolokoltsov AA, Davey RA, Ireton K. Gavicherla B, et al. Infect Immun. 2010 Nov;78(11):4532-41. doi: 10.1128/IAI.00802-10. Epub 2010 Sep 7. Infect Immun. 2010. PMID: 20823205 Free PMC article. - Role of Host Type IA Phosphoinositide 3-Kinase Pathway Components in Invasin-Mediated Internalization of Yersinia enterocolitica.
Dowd GC, Bhalla M, Kean B, Thomas R, Ireton K. Dowd GC, et al. Infect Immun. 2016 May 24;84(6):1826-1841. doi: 10.1128/IAI.00142-16. Print 2016 Jun. Infect Immun. 2016. PMID: 27068087 Free PMC article. - Entry of the bacterial pathogen Listeria monocytogenes into mammalian cells.
Ireton K. Ireton K. Cell Microbiol. 2007 Jun;9(6):1365-75. doi: 10.1111/j.1462-5822.2007.00933.x. Epub 2007 Apr 5. Cell Microbiol. 2007. PMID: 17419717 Review. - Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view.
Pizarro-Cerdá J, Kühbacher A, Cossart P. Pizarro-Cerdá J, et al. Cold Spring Harb Perspect Med. 2012 Nov 1;2(11):a010009. doi: 10.1101/cshperspect.a010009. Cold Spring Harb Perspect Med. 2012. PMID: 23125201 Free PMC article. Review.
Cited by
- Relative Roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes Uptake by Host Cells.
Phelps CC, Vadia S, Arnett E, Tan Y, Zhang X, Pathak-Sharma S, Gavrilin MA, Seveau S. Phelps CC, et al. Infect Immun. 2018 Sep 21;86(10):e00555-18. doi: 10.1128/IAI.00555-18. Print 2018 Oct. Infect Immun. 2018. PMID: 30061379 Free PMC article. - c-KIT signaling is targeted by pathogenic Yersinia to suppress the host immune response.
Micheva-Viteva SN, Shou Y, Nowak-Lovato KL, Rector KD, Hong-Geller E. Micheva-Viteva SN, et al. BMC Microbiol. 2013 Nov 9;13:249. doi: 10.1186/1471-2180-13-249. BMC Microbiol. 2013. PMID: 24206648 Free PMC article. - Host Serine/Threonine Kinases mTOR and Protein Kinase C-α Promote InlB-Mediated Entry of Listeria monocytogenes.
Bhalla M, Law D, Dowd GC, Ireton K. Bhalla M, et al. Infect Immun. 2017 Jun 20;85(7):e00087-17. doi: 10.1128/IAI.00087-17. Print 2017 Jul. Infect Immun. 2017. PMID: 28461391 Free PMC article. - _Listeria monocytogenes_-How This Pathogen Uses Its Virulence Mechanisms to Infect the Hosts.
Osek J, Wieczorek K. Osek J, et al. Pathogens. 2022 Dec 7;11(12):1491. doi: 10.3390/pathogens11121491. Pathogens. 2022. PMID: 36558825 Free PMC article. Review. - The Host GTPase Arf1 and Its Effectors AP1 and PICK1 Stimulate Actin Polymerization and Exocytosis To Promote Entry of Listeria monocytogenes.
Saila S, Gyanwali GC, Hussain M, Gianfelice A, Ireton K. Saila S, et al. Infect Immun. 2020 Jan 22;88(2):e00578-19. doi: 10.1128/IAI.00578-19. Print 2020 Jan 22. Infect Immun. 2020. PMID: 31740529 Free PMC article.
References
- Allam A, Niiro H, Clark EA, Marshall AJ. 2004. The adaptor protein Bam32 regulates Rac1 activation and actin remodeling through a phosphorylation-dependent mechanism. J. Biol. Chem. 279:39775–39782 - PubMed
- Bayascas JR. 2010. PDK1: the major transducer of PI 3-kinase actions. Curr. Top. Microbiol. Immunol. 346:9–29 - PubMed
- Bierne H, et al. 2005. WASP-related proteins, Abi and Ena/VASP are required for Listeria invasion induced by the Met receptor. J. Cell Sci. 118:1537–1547 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous