Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line - PubMed (original) (raw)
Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line
Sidney H Wang et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Transposon control is a critical process during reproduction. The PIWI family proteins can play a key role, using a piRNA-mediated slicing mechanism to suppress transposon activity posttranscriptionally. In Drosophila melanogaster, Piwi is predominantly localized in the nucleus and has been implicated in heterochromatin formation. Here, we use female germ-line-specific depletion to study Piwi function. This depletion of Piwi leads to infertility and to axis specification defects in the developing egg chambers; correspondingly, widespread loss of transposon silencing is observed. Germ-line Piwi does not appear to be required for piRNA production. Instead, Piwi requires Aubergine (and presumably secondary piRNA) for proper localization. A subset of transposons that show significant overexpression in germ-line Piwi-depleted ovaries exhibit a corresponding loss of HP1a and H3K9me2. Germ-line HP1a depletion also leads to a loss of transposon silencing, demonstrating the functional requirement for HP1a enrichment at these loci. Considering our results and those of others, we infer that germ-line Piwi functions downstream of piRNA production to promote silencing of some transposons via recruitment of HP1a. Thus, in addition to its better-known function in posttranscriptional silencing, piRNA also appears to function in a targeting mechanism for heterochromatin formation mediated by Piwi.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
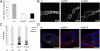
Germ-line–specific Piwi depletion leads to axis specification defects in developing egg chambers. (A) Quantitative RT-PCR analysis of the piwi expression level in germ-line Piwi knockdown ovaries. Expression levels are given relative to the RPL32 locus. (Bars represent the mean ± SEM) (B) Piwi antibody staining of developing egg chambers. Piwi depletion specifically in the germ line (red arrows) is achieved with either of two independent knockdown constructs without affecting the surrounding somatic follicle cells. (Full genotypes of piwiKD1 and piwiKD2 are given in
SI Appendix, Table S5
.) (C) The cumulative percentage of dorsal appendage phenotype of embryos produced by germ-line Piwi knockdown females. (N represents the total number of embryos scored for each genotype.) (D) Gurken (green) immunofluorescent staining of stage 9 developing egg chambers. The oocyte nucleus is indicated (asterisk). DAPI staining (blue) marks the nuclei and the actin filament (red) marks the cell boundaries. Gurken localization is diminished in the Piwi knockdown lines.
Fig. 2.
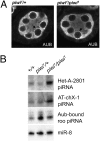
Depletion of germ-line Piwi does not disrupt Aub function. (A) Aub immunofluorescent staining of stage 4/5 egg chambers bearing piwi1/+ or piwi1/piwi1 germ line. The peri-nuclear structure nuage (black arrow) and Aub localization are not perturbed in the _piwi1/piwi_1 germ line. (B) Small RNA Northern blot analysis using three different piRNA probes, HeT-A-2801, AT-chX-1, and Aub-bound roo, along with a microRNA probe, miR-8, as a loading control. (A and B) Genotypes indicated are germ-line genotype at the piwi locus.
Fig. 3.
Germ-line Aub knockdown perturbs proper Piwi nuclear localization and leads to overexpression of some transposons. (A) Western blot analysis of Piwi or Aub protein levels in Aub knockdown ovaries shows no significant loss of Piwi. Myosin VI is used as the loading control; the volume of lysate loaded in each lane is indicated beneath. (B) Quantitative RT-PCR analysis of transposon expression levels in germ-line Aub knockdown ovaries. Expression levels are given relative to the RPL32 locus. (Bars represent the mean ± SEM.) (C) Piwi immunofluorescent staining of ovarioles. In the Aub knockdown germ line, Piwi is barely visible in the nuclei of early stage egg chambers (arrow) in contrast to wild type. (D) The diffuse pattern of Piwi staining in an Aub knockdown germ line is most apparent in stage 2/3 egg chambers. (E) Piwi immunofluorescent staining of stage 6/7 egg chambers. DNA staining is shown in red to delineate the nucleus. The overall Piwi signal in the Aub knockdown egg chambers is adjusted so that the signal strength in the germline nuclei matches the corresponding region in the wild-type egg chamber. (Scale bars: 5 μm.)
Fig. 4.
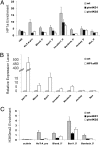
Germ-line Piwi functions in silencing some transposons through an HP1a-dependent chromatin-based mechanism. (A) ChIP-quantitative PCR analysis at 5′ ends or promoter regions (as indicated in the label) of a set of transposons using antibodies against HP1a in germ-line Piwi knockdown ovaries. The enrichment levels are relative to the α-actinin locus. (B) Quantitative RT-PCR analysis of expression levels for the same set of transposons in germ-line HP1a knockdown ovaries. Fold expression levels are relative to RPL32 expression. (C) ChIP-quantitative PCR analysis at 5′/promoter regions of a set of transposons using antibodies against H3K9me2 in germ-line Piwi knockdown ovaries. The enrichment levels are relative to the 18S ribosomal DNA locus. Bars represent mean ± SEM of three biological replicate experiments.
Similar articles
- Maternally inherited piRNAs direct transient heterochromatin formation at active transposons during early Drosophila embryogenesis.
Fabry MH, Falconio FA, Joud F, Lythgoe EK, Czech B, Hannon GJ. Fabry MH, et al. Elife. 2021 Jul 8;10:e68573. doi: 10.7554/eLife.68573. Elife. 2021. PMID: 34236313 Free PMC article. - piRNA-mediated regulation of transposon alternative splicing in the soma and germ line.
Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. Teixeira FK, et al. Nature. 2017 Dec 14;552(7684):268-272. doi: 10.1038/nature25018. Epub 2017 Dec 6. Nature. 2017. PMID: 29211718 Free PMC article. - Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery.
Sienski G, Batki J, Senti KA, Dönertas D, Tirian L, Meixner K, Brennecke J. Sienski G, et al. Genes Dev. 2015 Nov 1;29(21):2258-71. doi: 10.1101/gad.271908.115. Epub 2015 Oct 22. Genes Dev. 2015. PMID: 26494711 Free PMC article. - Untangling the web: the diverse functions of the PIWI/piRNA pathway.
Mani SR, Juliano CE. Mani SR, et al. Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review. - Two distinct transcriptional controls triggered by nuclear Piwi-piRISCs in the Drosophila piRNA pathway.
Sato K, Siomi MC. Sato K, et al. Curr Opin Struct Biol. 2018 Dec;53:69-76. doi: 10.1016/j.sbi.2018.06.005. Epub 2018 Jul 7. Curr Opin Struct Biol. 2018. PMID: 29990672 Review.
Cited by
- Functions of HP1 proteins in transcriptional regulation.
Schoelz JM, Riddle NC. Schoelz JM, et al. Epigenetics Chromatin. 2022 May 7;15(1):14. doi: 10.1186/s13072-022-00453-8. Epigenetics Chromatin. 2022. PMID: 35526078 Free PMC article. Review. - Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing.
Murano K, Iwasaki YW, Ishizu H, Mashiko A, Shibuya A, Kondo S, Adachi S, Suzuki S, Saito K, Natsume T, Siomi MC, Siomi H. Murano K, et al. EMBO J. 2019 Sep 2;38(17):e102870. doi: 10.15252/embj.2019102870. Epub 2019 Aug 1. EMBO J. 2019. PMID: 31368590 Free PMC article. - Complex Genetic Interactions between Piwi and HP1a in the Repression of Transposable Elements and Tissue-Specific Genes in the Ovarian Germline.
Ilyin AA, Stolyarenko AD, Zenkin N, Klenov MS. Ilyin AA, et al. Int J Mol Sci. 2021 Dec 14;22(24):13430. doi: 10.3390/ijms222413430. Int J Mol Sci. 2021. PMID: 34948223 Free PMC article. - Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline.
Radion E, Morgunova V, Ryazansky S, Akulenko N, Lavrov S, Abramov Y, Komarov PA, Glukhov SI, Olovnikov I, Kalmykova A. Radion E, et al. Epigenetics Chromatin. 2018 Jul 12;11(1):40. doi: 10.1186/s13072-018-0210-4. Epigenetics Chromatin. 2018. PMID: 30001204 Free PMC article. - Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, Patel D, Aravin AA. Le Thomas A, et al. Genes Dev. 2014 Aug 1;28(15):1667-80. doi: 10.1101/gad.245514.114. Genes Dev. 2014. PMID: 25085419 Free PMC article.
References
- Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases