Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction - PubMed (original) (raw)
Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction
James D Orth et al. Mol Biol Cell. 2012 Feb.
Abstract
Mitotic arrest induced by antimitotic drugs can cause apoptosis or p53-dependent cell cycle arrest. It can also cause DNA damage, but the relationship between these events has been unclear. Live, single-cell imaging in human cancer cells responding to an antimitotic kinesin-5 inhibitor and additional antimitotic drugs revealed strong induction of p53 after cells slipped from prolonged mitotic arrest into G1. We investigated the cause of this induction. We detected DNA damage late in mitotic arrest and also after slippage. This damage was inhibited by treatment with caspase inhibitors and by stable expression of mutant, noncleavable inhibitor of caspase-activated DNase, which prevents activation of the apoptosis-associated nuclease caspase-activated DNase (CAD). These treatments also inhibited induction of p53 after slippage from prolonged arrest. DNA damage was not due to full apoptosis, since most cytochrome C was still sequestered in mitochondria when damage occurred. We conclude that prolonged mitotic arrest partially activates the apoptotic pathway. This partly activates CAD, causing limited DNA damage and p53 induction after slippage. Increased DNA damage via caspases and CAD may be an important aspect of antimitotic drug action. More speculatively, partial activation of CAD may explain the DNA-damaging effects of diverse cellular stresses that do not immediately trigger apoptosis.
Figures
FIGURE 1:
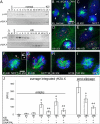
Prolonged mitotic arrest results in two types of postslippage p53 induction dynamics. (A) Mock, untreated (M), G2-synchronized cells (synch; see Materials and Methods), and synchronized cells released into normal (n) or K5I-medium (K5I) were immunoblotted for p53, p21, and the mitotic marker p-H3. p53 and p21 were not persistently induced to high levels in normal medium. Release into K5I resulted in p53 induction beginning at 12–16 h and p21 induction after 16 h. All blots in each panel are from the same gels and/or samples. (B–D) Live MCF7 cells expressing p53-Venus were imaged for >60 h. Times are from start of time lapse. First frame is start of mitotic arrest. No p53 increase was noted during mitotic arrest. Sixty percent of arrested cells showed sustained, high-level, p53-induction (cell A), while 30% showed a pulsatile response with a large first pulse (cell B). (B and C) Scale bars: 10 μm. See Movies S1 and S2 and note additional cells in the same microscopic field. Vertical lines in representative traces (D) mark mitotic slippage. RFU, relative fluorescence units. (E) Box-and-whisker plots of peak p53 levels in individual cells with sustained and pulsatile dynamics; p53 induction is significantly higher in cells with sustained induction. For cells in each group, the peak p53 intensity is determined, and the resulting values are assorted in ascending order. The boxes represent the second and third quartile of the population, the red horizontal line represents the median. Upper and lower “whiskers” represent data points from the first and fourth quartile that are within 1.5-fold of the interquartile range. Outliers are marked by red +. The notches about the median represent the 95% confidence interval. As the notches between the two compared populations do not overlap, the median values are significantly different (p < 0.0001 for sustained vs. pulsatile). (F) Box-and-whisker plots of hours arrested for the types of p53 dynamics. The medians are not significantly different. Forty cells were tracked: 26 showed a sustained induction, and 14 were pulsatile.
FIGURE 2:
DNA damage after K5I initially increases late in mitotic arrest and involves DNA damage–response kinases, ATM and DNA-PK. (A) Mock, untreated (M), G2-synchronized, and cells released into normal (n) or K5I medium. Normal shows low-level γH2A.X, but K5I shows significantly higher levels beginning at mitotic slippage (decreased p-H3). All blots in each panel are from the same gels and/or samples. (B–E and J) Green, microtubules (mt); red, γH2A.X; blue, DNA. Only γH2A.X on the mitotic chromosomes or interphase DNA was quantified. n, nucleus; arrows, mitotic cells; RFU, relative fluorescence units. (C) Scale bar: 15 μm (applies also to B–E). Normal mitotic cells and mitosis-arrested cells at 4 and 8 h K5I show low γH2A.X (red), but 16 h K5I-treated cells shows high levels of γH2A.X. In 16 h arrested cells, γH2A.X is blocked by ATM inhibition (ATMi) but not by DNA-PK inhibition (DNA-PKi); combined ATMi and DNA-PKi at 16 h was similar to ATMi results. Postslippage γH2A.X at 48 h is high and was dependent on both ATM and DNA-PK. Values in (J) are average integrated fluorescence intensity per cell (± SE). At 4 h with no K5I, ATMi, or DNA-PKi, n = 15 cells, two experiments. All other conditions, n > 100 cells, ≥ two experiments. Mitotic conditions: *, p < 0.05 vs. 4 h control; **, p < 0.05 vs. 16 h K5I or 16 h K5I + DNA-PKi. Postmitotic conditions: *, p < 0.05 vs. 48 h K5I; **, p < 0.05 48 h K5I or 48 h K5I + ATMi. (F–I) γH2A.X occurs during mitotic arrest in different cell lines, is not p53-dependent (MCF7 sh p53), and occurs in cancer and noncancer cells (RPE1). Inset in (F) shows separate chromosome (top) and γH2A.X (bottom) channels for the γH2A.X foci (arrow). (F) Scale bar: 5 μm (applies also to G–I).
FIGURE 3:
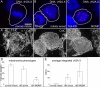
DNA damage during mitotic arrest occurs without full mitochondrial MOMP. (A–C′) DNA, γH2A.X and CytC staining in the same mitotic cells. (D) Quantification of the SD of the average CytC fluorescence pixel intensity in individual mitotic or MOMP cells (encircled) shows that 16 h mitosis-arrested cells (B′) are morphologically altered compared with normal (A′), and have a lower average CytC SD than normal mitotic cells, but a higher value than MOMP cells (C′). (E) Normal mitotic cells (A) have very low γH2A.X compared with 16 h arrest (B). MOMP cells (C) on average showed more DNA damage, but this was highly variable and not statistically significant in this small population (t test 16 h MOMP vs. 16 h arrest, 0.33). Average values are reported (± SE). (A) Scale bar: 5 μm (applies to all cell images). Control, n = 68; 16 h mitotic, n = 295; 16 h MOMP, n = 16; two experiments each condition. (D) *, p < 0.05 vs. control mitotic; **, p < 0.05 vs. 16 h mitotic and normal mitotic. (E) *, p < 0.05 vs. normal mitotic.
FIGURE 4:
DNA damage during mitotic arrest is dependent upon caspase-9 and -7. (A and B) Green, microtubules (mt); red, γH2A.X; blue, DNA. G2-synchronized cells released for 16 h into K5I or K5I with the caspase inhibitor zVAD-fmk show γH2A.X increase is caspase-dependent. Scale bar: 5 μm. (C) γH2A.X quantification of single cells shows caspase-dependent DNA damage in 16 h mitosis-arrested cells and 48 h postslippage cells. Average integrated fluorescence intensity (± SE). RFU, relative fluorescence units. n = 50+ for each condition; two experiments . *, p < 0.05 vs. 16 h K5I; **, p < 0.05 vs. 48 h K5I. (D) Mock, untreated (M) and G2-synchronized (synch) cells, and synchronized cells released into normal medium show no activation of caspase-9 or -7, as measured by loss of full-length caspase-9 (Casp9) or increase in cleaved caspase-7 (c Casp7). γH2A.X also did not increase compared with 24 h K5I. (E) Release into K5I shows loss of caspase-9, increased cleaved caspase-7, and increased γH2A.X in late arrest/early slippage (MPM2 blot) blocked by zVAD-fmk. MPM2 indicates several bands during mitosis. (F) Caspase-9 inhibition with zLEHD-fmk prevented caspase-9 and -7 activation (c Casp9 and c Casp7) and inhibited γH2A.X increase. (G) Caspase-7 inhibition with zDEVD-fmk did not prevent caspase-9 activation, but largely blocked caspase-7, resulting in decreased γH2A.X. Note: the slightly higher-migrating cleaved caspase-7 band in the presence of caspase inhibitors in (E, F, and G) represents incomplete processing (activation). All blots in each panel are from the same gels and/or samples. Dynein and actin are the loading controls.
FIGURE 5:
Mitotic DNA damage occurs downstream of caspase-dependent, partial cleavage of ICAD. (A) Immunoprecipitation (IP) of ICAD followed by blotting for cleaved ICAD (c ICAD) at 24 h K5I showed less cleavage than 24 h 1 μM STS but more than UV treatment (see Materials and Methods). Ten percent input blot: ICAD shows total ICAD levels. (B) Mock, untreated (M) and G2-synchronized (synch) cells, and cells released into K5I showed decreased ICAD from 8 to 48 h and increased c ICAD from 16 h, correlating well with late mitotic arrest/slippage (p-H3). Dynein is the loading control. (C) Immunoprecipitation of ICAD followed by blotting for c ICAD showed partial cleavage of ICAD at 24 h K5I treatment is blocked by zVAD-fmk or expression of noncleavable ICAD (clone A). All blots in each panel are from the same gels and/or samples. (D) Noncleavable ICAD-expressing clone A and B showed the same background levels of γH2A.X as MCF7 CELLS. G2-synchronized clone A and B released into K5I for 16 h showed severely inhibited γH2A.X compared with MCF7 cells. Average integrated fluorescence intensity (± SE). RFU, relative fluorescence units. No K5I: MCF7, n = 31; clone A, n = 12; clone B, n = 16. 16 h K5I: MCF7, n = 210; clone A, n = 166; clone B, n = 142; two experiments each condition. *, p < 0.05 vs. MCF7 + 16 h K5I.
FIGURE 6:
p53 induction after mitotic DNA damage is caspase- and ICAD/CAD-dependent. (A) Immunoblotting of mock, untreated (M) and G2-synchronized (synch) cells, and cells released into K5I showed strong, persistent induction of p53 after release that was significantly blocked by caspase-9 or -7 inhibition. Actin and dynein are loading controls. (B) Live MCF7 p53-Venus cells. First vertical green line in traces is entry into mitotic arrest, second is mitotic slippage. In K5I, p53 is often induced to sustained, high levels. Cells cotreated K5I and zVAD-fmk showed only transient induction of p53 to low levels. RFU, relative fluorescence units. Caspase inhibition resulted in decreased fold induction (average of all cells ± SE) in individual cells and in fewer cells with >2 fold induction. Fold induction is the ratio of the peak intensity to the lowest intensity in each cell. K5I, n = 32; K5I + zVAD-fmk, n = 49. (C) Mock, untreated (M), MCF7, and noncleavable ICAD clone B were G2-synchronized (synch) and released into K5I. When ICAD cleavage is blocked, both p53 induction and DNA damage (γH2A.X) are decreased. Importantly, caspase-7 remains activated after K5I treatment. Dynein is loading control. All blots in each panel are from the same gels and/or samples. (D) Summary of pathway causing DNA damage during mitotic arrest that results in postslippage p53 induction.
Similar articles
- Paclitaxel-induced aberrant mitosis and mitotic slippage efficiently lead to proliferative death irrespective of canonical apoptosis and p53.
Yasuhira S, Shibazaki M, Nishiya M, Maesawa C. Yasuhira S, et al. Cell Cycle. 2016 Dec;15(23):3268-3277. doi: 10.1080/15384101.2016.1242537. Epub 2016 Oct 20. Cell Cycle. 2016. PMID: 27764550 Free PMC article. - Tumor-Associated Macrophages Suppress the Cytotoxic Activity of Antimitotic Agents.
Olson OC, Kim H, Quail DF, Foley EA, Joyce JA. Olson OC, et al. Cell Rep. 2017 Apr 4;19(1):101-113. doi: 10.1016/j.celrep.2017.03.038. Cell Rep. 2017. PMID: 28380350 Free PMC article. - Cellular responses to a prolonged delay in mitosis are determined by a DNA damage response controlled by Bcl-2 family proteins.
Colin DJ, Hain KO, Allan LA, Clarke PR. Colin DJ, et al. Open Biol. 2015 Mar;5(3):140156. doi: 10.1098/rsob.140156. Open Biol. 2015. PMID: 25761368 Free PMC article. - Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events.
Blagosklonny MV. Blagosklonny MV. Cell Cycle. 2007 Jan 1;6(1):70-4. doi: 10.4161/cc.6.1.3682. Epub 2007 Jan 9. Cell Cycle. 2007. PMID: 17245109 Review. - The role of p53 in the response to mitotic spindle damage.
Meek DW. Meek DW. Pathol Biol (Paris). 2000 Apr;48(3):246-54. Pathol Biol (Paris). 2000. PMID: 10858957 Review.
Cited by
- Short- and long-term effects of chromosome mis-segregation and aneuploidy.
Santaguida S, Amon A. Santaguida S, et al. Nat Rev Mol Cell Biol. 2015 Aug;16(8):473-85. doi: 10.1038/nrm4025. Nat Rev Mol Cell Biol. 2015. PMID: 26204159 Review. - Generation and Purification of Tetraploid Cells.
Shenk EM, Ganem NJ. Shenk EM, et al. Methods Mol Biol. 2016;1413:393-401. doi: 10.1007/978-1-4939-3542-0_24. Methods Mol Biol. 2016. PMID: 27193862 Free PMC article. - Homeostatic control of polo-like kinase-1 engenders non-genetic heterogeneity in G2 checkpoint fidelity and timing.
Liang H, Esposito A, De S, Ber S, Collin P, Surana U, Venkitaraman AR. Liang H, et al. Nat Commun. 2014 Jun 4;5:4048. doi: 10.1038/ncomms5048. Nat Commun. 2014. PMID: 24893992 Free PMC article. - Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival.
Elsing AN, Aspelin C, Björk JK, Bergman HA, Himanen SV, Kallio MJ, Roos-Mattjus P, Sistonen L. Elsing AN, et al. J Cell Biol. 2014 Sep 15;206(6):735-49. doi: 10.1083/jcb.201402002. Epub 2014 Sep 8. J Cell Biol. 2014. PMID: 25202032 Free PMC article. - Emergence of spatio-temporal variations in chemotherapeutic drug efficacy: in-vitro and in-Silico 3D tumour spheroid studies.
Sheraton MV, Chiew GGY, Melnikov V, Tan EY, Luo KQ, Verma N, Sloot PMA. Sheraton MV, et al. BMC Cancer. 2020 Dec 7;20(1):1201. doi: 10.1186/s12885-020-07677-5. BMC Cancer. 2020. PMID: 33287759 Free PMC article.
References
- Bacus SS, Gudkov AV, Lowe M, Lyass L, Yung Y, Komarov AP, Keyomarsi K, Yarden Y, Seger R. Taxol-induced apoptosis depends on MAP kinase pathways (ERK and p38) and is independent of p53. Oncogene. 2001;20:147–155. - PubMed
- Barboule N, Chadebech P, Baldin V, Vidal S, Valette A. Involvement of p21 in mitotic exit after paclitaxel treatment in MCF-7 breast adenocarcinoma cell line. Oncogene. 1997;15:2867–2875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA164448/CA/NCI NIH HHS/United States
- CA139980/CA/NCI NIH HHS/United States
- P01 CA139980/CA/NCI NIH HHS/United States
- GM083303/GM/NIGMS NIH HHS/United States
- R01 GM083303/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous