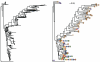Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data - PubMed (original) (raw)
Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data
Paul J McMurdie et al. Pac Symp Biocomput. 2012.
Abstract
We present a detailed description of a new Bioconductor package, phyloseq, for integrated data and analysis of taxonomically-clustered phylogenetic sequencing data in conjunction with related data types. The phyloseq package integrates abundance data, phylogenetic information and covariates so that exploratory transformations, plots, and confirmatory testing and diagnostic plots can be carried out seamlessly. The package is built following the S4 object-oriented framework of the R language so that once the data have been input the user can easily transform, plot and analyze the data. We present some examples that highlight the methods and the ease with which we can leverage existing packages.
Figures
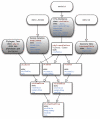
Fig. 1
Classes and inheritance in the phyloseq package. Core data classes are shown with grey fill and rounded corners. The class name and its slots are shown with red- or blue-shaded text, respectively. Inheritance is indicated graphically by arrows. Lines without arrows indicate that a higher-order object contains a slot with the associated class as one of its components.
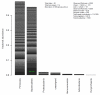
Fig. 2
Example of a default plot method for summarizing an object of class otuSam-Tax. Each phyloseq class has a specialized plot method for summarizing its data. In this case, relative abundance is shown quantitatively in a stacked barplot by phylum. Different taxa within a stack are differentiated by an alternating series of grayscale. The OTU identifier of taxa comprising a large enough fraction of the total community, 5% in this case, is labeled on the corresponding bar segment. Several diversity/richness indices are also shown.
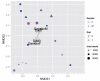
Fig. 3
NMDS ordination graphic generated by wunifracMDS. The NMDS coordinates are generated by metaMDS(), with the weighted-UniFrac distance matrix as argument, and 2-dimensions specified by default. A separate analysis was done using adonis(), which also did not find a compelling association between the weighted UniFrac distances and the gender (p = 0.29) or diet (p = 0.9) of subjects in the study.
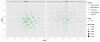
Fig. 4
Redundancy analysis and Constrained Correspondence Analysis. (Left) Redundancy analysis applied to a thresholded, ranked-transformed abundance table that had been trimmed such that only the phyla accounting for the top 99% of taxa are included. (Right) Original trimmed abundance table (no transformation nor threshold) subjected to Constrained Correspondence Analysis (CCA), constrained on a subject’s diet and gender.
Fig. 5
Enlarged RDA and CCA plots emphasizing the taxa (species) coordinates. Graphics were produced with calcplotrda() or calcplotcca() convenience wrappers in phyloseq, which utilize analysis and graphics tools from the vegan and ggplot2 packages, respectively. Only the phyla accounting for the top 99% of taxa are included.
Fig. 6
Example of phylogenetic sequence data before and after basic clustering with tipglom() function. (Left) Standard phylogram produced using default plotting function and no OTU clustering. (Right) Annotated phylogram after OTU clustering with tipglom(). Different symbols next to each tip indicate different samples in which the OTU was observed. The number inside each symbol indicates the respective number of individuals of a given OTU were observed in each sample.
Similar articles
- REPRODUCIBLE RESEARCH WORKFLOW IN R FOR THE ANALYSIS OF PERSONALIZED HUMAN MICROBIOME DATA.
Callahan B, Proctor D, Relman D, Fukuyama J, Holmes S. Callahan B, et al. Pac Symp Biocomput. 2016;21:183-94. Pac Symp Biocomput. 2016. PMID: 26776185 Free PMC article. - phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data.
McMurdie PJ, Holmes S. McMurdie PJ, et al. PLoS One. 2013 Apr 22;8(4):e61217. doi: 10.1371/journal.pone.0061217. Print 2013. PLoS One. 2013. PMID: 23630581 Free PMC article. - Advancing our understanding of the human microbiome using QIIME.
Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG, Knight R. Navas-Molina JA, et al. Methods Enzymol. 2013;531:371-444. doi: 10.1016/B978-0-12-407863-5.00019-8. Methods Enzymol. 2013. PMID: 24060131 Free PMC article. - Using R and Bioconductor in Clinical Genomics and Transcriptomics.
Sepulveda JL. Sepulveda JL. J Mol Diagn. 2020 Jan;22(1):3-20. doi: 10.1016/j.jmoldx.2019.08.006. Epub 2019 Oct 9. J Mol Diagn. 2020. PMID: 31605800 Review. - Orchestrating high-throughput genomic analysis with Bioconductor.
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Huber W, et al. Nat Methods. 2015 Feb;12(2):115-21. doi: 10.1038/nmeth.3252. Nat Methods. 2015. PMID: 25633503 Free PMC article. Review.
Cited by
- Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum.
LeMay-Nedjelski L, Butcher J, Ley SH, Asbury MR, Hanley AJ, Kiss A, Unger S, Copeland JK, Wang PW, Zinman B, Stintzi A, O'Connor DL. LeMay-Nedjelski L, et al. BMC Microbiol. 2020 Jul 20;20(1):219. doi: 10.1186/s12866-020-01901-9. BMC Microbiol. 2020. PMID: 32689933 Free PMC article. Clinical Trial. - High-performing cross-dataset machine learning reveals robust microbiota alteration in secondary apical periodontitis.
Li H, Li J, Hu J, Chen J, Zhou W. Li H, et al. Front Cell Infect Microbiol. 2024 Jun 21;14:1393108. doi: 10.3389/fcimb.2024.1393108. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38975327 Free PMC article. - Urogenital schistosomiasis is associated with signatures of microbiome dysbiosis in Nigerian adolescents.
Ajibola O, Rowan AD, Ogedengbe CO, Mshelia MB, Cabral DJ, Eze AA, Obaro S, Belenky P. Ajibola O, et al. Sci Rep. 2019 Jan 29;9(1):829. doi: 10.1038/s41598-018-36709-1. Sci Rep. 2019. PMID: 30696838 Free PMC article. - Unraveling the effects of the gut microbiota composition and function on horse endurance physiology.
Plancade S, Clark A, Philippe C, Helbling JC, Moisan MP, Esquerré D, Le Moyec L, Robert C, Barrey E, Mach N. Plancade S, et al. Sci Rep. 2019 Jul 3;9(1):9620. doi: 10.1038/s41598-019-46118-7. Sci Rep. 2019. PMID: 31270376 Free PMC article. - Functional genes to assess nitrogen cycling and aromatic hydrocarbon degradation: primers and processing matter.
Penton CR, Johnson TA, Quensen JF 3rd, Iwai S, Cole JR, Tiedje JM. Penton CR, et al. Front Microbiol. 2013 Sep 17;4:279. doi: 10.3389/fmicb.2013.00279. eCollection 2013. Front Microbiol. 2013. PMID: 24062736 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources