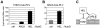A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication - PubMed (original) (raw)
A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication
Masaki Nishikiori et al. PLoS Pathog. 2011 Dec.
Abstract
Tomato mosaic virus (ToMV), like other eukaryotic positive-strand RNA viruses, replicates its genomic RNA in replication complexes formed on intracellular membranes. Previous studies showed that a host seven-pass transmembrane protein TOM1 is necessary for efficient ToMV multiplication. Here, we show that a small GTP-binding protein ARL8, along with TOM1, is co-purified with a FLAG epitope-tagged ToMV 180K replication protein from solubilized membranes of ToMV-infected tobacco (Nicotiana tabacum) cells. When solubilized membranes of ToMV-infected tobacco cells that expressed FLAG-tagged ARL8 were subjected to immunopurification with anti-FLAG antibody, ToMV 130K and 180K replication proteins and TOM1 were co-purified and the purified fraction showed RNA-dependent RNA polymerase activity that transcribed ToMV RNA. From uninfected cells, TOM1 co-purified with FLAG-tagged ARL8 less efficiently, suggesting that a complex containing ToMV replication proteins, TOM1, and ARL8 are formed on membranes in infected cells. In Arabidopsis thaliana, ARL8 consists of four family members. Simultaneous mutations in two specific ARL8 genes completely inhibited tobamovirus multiplication. In an in vitro ToMV RNA translation-replication system, the lack of either TOM1 or ARL8 proteins inhibited the production of replicative-form RNA, indicating that TOM1 and ARL8 are required for efficient negative-strand RNA synthesis. When ToMV 130K protein was co-expressed with TOM1 and ARL8 in yeast, RNA 5'-capping activity was detected in the membrane fraction. This activity was undetectable or very weak when the 130K protein was expressed alone or with either TOM1 or ARL8. Taken together, these results suggest that TOM1 and ARL8 are components of ToMV RNA replication complexes and play crucial roles in a process toward activation of the replication proteins' RNA synthesizing and capping functions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
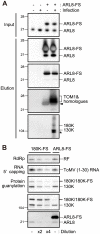
Figure 1. Analysis of proteins co-purified with the 180K-FS protein.
(A) Schematic representation of ToMV, ToMV-GFP, and ToMV-180FS-GFP. (B) Silver staining of proteins contained in the FLAG-purified fractions “Copyright © American Society for Microbiology, Journal of Virology, 80, 2006, 8459-8468, doi:10.1128/JVI.00545-06”. Soluble and membrane fractions of ToMV-GFP- or ToMV-180FS-GFP-infected and evacuolated tobacco BY-2 protoplasts were prepared, treated with LPC, clarified by centrifugation, and subjected to affinity purification with monoclonal anti-FLAG antibody. Co-purified proteins were separated by SDS-12% PAGE and silver-stained (upper panel). The positions of protein markers are shown on the left with their molecular weights (x10−3). Proteins identified by LC-MS/MS are shown on the right. The bottom panel shows an immunoblot of the fractions detected with anti-ARL8 antibodies. (C) Alignment of deduced amino acid sequences of A. thaliana (At) and N. tabacum (Nt) ARL8 proteins. The sequence data of the AtARL8 proteins were obtained from the Arabidopsis Information Resource (
). Peptides identified by LC-MS/MS are indicated by red letters. (D) Phylogenetic tree of ARF family proteins. An unrooted tree was constructed using ClustalW with standard parameters and drawn using TreeView. Scale bar represents 0.1-amino acid substitutions per site. Abbreviations: At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens.
Figure 2. Affinity purification of ARL8-FS from uninfected and ToMV-infected BY-2 cells.
(A) Copurification of TOM1, 130K and 180K proteins with NtARL8a-FS. The P15 membrane fractions were prepared from the extracts of NtARL8a-FS-expressing BY-2 cells that had or had not been infected with ToMV-GFP. NtARL8a-FS was solubilized with LPC and immunopurified with the anti-FLAG antibody . NtARL8a-FS and endogenous ARL8 proteins were detected by immunoblotting and Coomassie brilliant blue-staining of the blotted membrane (second and third panels from the top, respectively). TOM1 and ToMV 130K and 180K replication proteins were detected by immunoblotting. Control experiments with BY-2 cells that did not express NtARL8a-FS were performed in parallel. The arrowhead shows signals corresponding to the crossreaction of anti-TOM1 antibodies with NtARL8a-FS. The asterisk indicates a degradation product of the 130K/180K proteins. The positions of protein markers are shown on the left with their molecular weights (x10−3). (B) Copurification of ToMV RdRp and capping activities with ARL8-FS. The P15 membrane fractions were prepared from ARL8-FS-expressing and ToMV-GFP-infected BY-2 cells or from ToMV-180FS-GFP-infected BY-2 cells that were evacuolated. The fractions were solubilized with LPC and immunopurified with the anti-FLAG antibody . The FLAG-purified fractions were subjected to RdRp, RNA 5′ capping, and protein guanylation assays. The fractions were also subjected to immunoblot analysis to detect ToMV replication proteins and ARL8. Where specified, the fraction was diluted 2- or 4-fold. The RdRp reaction was performed in the presence of [α-32P]CTP using exogenously added ToMV RNA as a template as described previously , and analyzed by PAGE. RNA 5′ capping and protein guanylation reactions were performed and products were analyzed as described in the Materials and Methods. 32P-labeled bands were detected with an image analyzer (BAS 2500, Fujifilm). The positions corresponding to double-stranded ToMV RNA (RF), ToMV (1–30) RNA and 130K, 180K (180K-FS), and ARL8 (ARL8-FS) are indicated on the right.
Figure 3. Interactions between TOM1, ARL8, and the helicase domain polypeptide (HEL) of ToMV replication proteins.
(A) Split-ubiquitin assay using TOM1-Cub-PLV-expressing yeast. The graph shows β-galactosidase activity in the yeast reporter strain L40 coexpressing TOM1-Cub-PLV and either NubG-ALG5 (ctrl) , ARL8-NubG (ARL8), or NubG-HEL (HEL). ALG5 is an unrelated yeast protein used as a negative control. (B) Split-ubiquitin assay using ARL8-Cub-PLV-expressing yeast. The graph shows β-galactosidase activity in L40 yeast coexpressing ARL8-Cub-PLV and either NubG-ALG5 (ctrl) or NubG-HEL (HEL). Averages and standard deviations of β-galactosidase activity (Miller units) for three or four independent yeast transformants are indicated in panels A and B. (C) A model of interaction between TOM1, ARL8, and ToMV HEL polypeptide.
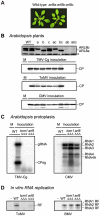
Figure 4. Effects of single and multiple mutations in A. thaliana ARL8 family members on virus multiplication.
(A) Wild-type (WT) and arl8 triple mutant plants. The photograph was taken 5 weeks after sowing. (B) Effects of arl8 mutations on the multiplication of TMV-Cg, ToMV, and CMV. Wild-type and arl8a, arl8b, and/or arl8c single, double, and triple mutant A. thaliana plants were inoculated with TMV-Cg, ToMV or CMV, as described previously . Inoculated leaves were harvested 2 days after inoculation (for TMV-Cg and CMV) or 7 days after inoculation (for ToMV), and the CPs were detected by immunoblotting using a 13% acrylamide, 0.35% bisacrylamide gel. ARL8a and ARL8b proteins were also detected (top panel). The asterisk denotes a background signal derived from cross-reactivity of the anti-ARL8 antiserum to a cellular protein, which served as loading controls. M indicates mock inoculation. (C) Effects of tom1 tom3 thh1 and arl8a arl8b arl8 triple mutations on the multiplication of TMV-Cg (left panel) and CMV (right panel) in A. thaliana protoplasts. Protoplasts with each genotype were isolated from suspension-cultured cells, and inoculated with TMV-Cg and CMV RNA by electroporation . Total RNA was isolated from the protoplasts 20 h after inoculation, and the accumulation of TMV-Cg and CMV RNA was analyzed by Northern blotting as described previously . (D) Effect of the loss of TOM1 and ARL8 on the production of viral RF RNA in the in vitro translation-replication system. A ToMV RNA derivative TL180SF and BMV virion RNAs were translated in mdBYL and ToMV PMTC were purified by 100,000 x g centrifugation and further with an anti-FLAG antibody to avoid contamination of soluble ARL8. For BMV, ribonucleoprotein complexes were purified by 100,000 x g centrifugation. The purified ribonucleoprotein fractions were mixed with membranes derived from protoplasts of suspension-cultured wild-type, tom1 or arl8 triple mutant A. thaliana cells. After the RdRp reaction, RNA samples were prepared, treated with a single-strand-specific S1 nuclease, and then analyzed by PAGE and autoradiography.
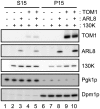
Figure 5. Effects of TOM1 and ARL8 coexpression on the membrane-association of ToMV 130K protein.
The soluble (S15) and membrane (P15) fractions were prepared from yeasts expressing the indicated proteins and subjected to immunoblot analysis using antibodies against TOM1, ARL8, ToMV 130K, a yeast cytoplasmic protein Pgk1p, and an ER-localized protein Dpm1p. S15 samples prepared from 0.12 OD600 units of yeast cells or P15 samples prepared from 0.6 OD600 units of yeast cells were applied in each lane.
Figure 6. Effects of TOM1 and ARL8 coexpression on RNA 5′ capping functions of ToMV 130K protein.
(A) A possible mechanism of RNA 5′ capping by tobamovirus replication proteins. The phosphate group at the α position of GTP is shown in red. The 5′ phosphate groups of the substrate RNA are shown in blue. See text for details. AdoHcy: _S_-Adenosyl-L-homocysteine. (B) Effects of the coexpression of TOM1 and ARL8 on RNA 5′ capping and guanylation of ToMV 130K protein expressed in yeast. P15 membrane fractions were prepared from the indicated yeast and BY-2 cells. The fractions were treated with LPC and subjected to immunopurification with anti-FLAG antibody . The protein samples were prepared so that the concentration of the replication proteins was similar (samples for lanes 1 and 6 were prepared and analyzed in the same way as for those for lanes 2 and 7, respectively). The samples were incubated at 25°C for 60 min with [α-32P]GTP, AdoMet and uncapped ToMV (1–30) RNA with 5′-triphosphate or 5′-diphosphate. After the reaction, RNA was purified by phenol extraction and separated by 8 M urea-9% PAGE. Fractions of the purified RNA samples were treated with TAP and analyzed similarly. 32P-labeled RNA was visualized with an image analyzer (BAS2500, Fujifilm). The protein samples were also subjected to immunoblot analysis with anti-ToMV replication protein antibodies and protein guanylation assay as described in the Materials and Methods.
Similar articles
- The resistance protein Tm-1 inhibits formation of a Tomato mosaic virus replication protein-host membrane protein complex.
Ishibashi K, Ishikawa M. Ishibashi K, et al. J Virol. 2013 Jul;87(14):7933-9. doi: 10.1128/JVI.00743-13. Epub 2013 May 8. J Virol. 2013. PMID: 23658455 Free PMC article. - Overexpression of a host factor TOM1 inhibits tomato mosaic virus propagation and suppression of RNA silencing.
Hagiwara-Komoda Y, Hirai K, Mochizuki A, Nishiguchi M, Meshi T, Ishikawa M. Hagiwara-Komoda Y, et al. Virology. 2008 Jun 20;376(1):132-9. doi: 10.1016/j.virol.2008.03.020. Epub 2008 Apr 28. Virology. 2008. PMID: 18440043 - Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound.
Nishikiori M, Dohi K, Mori M, Meshi T, Naito S, Ishikawa M. Nishikiori M, et al. J Virol. 2006 Sep;80(17):8459-68. doi: 10.1128/JVI.00545-06. J Virol. 2006. PMID: 16912296 Free PMC article. - Replication of Tobamovirus RNA.
Ishibashi K, Ishikawa M. Ishibashi K, et al. Annu Rev Phytopathol. 2016 Aug 4;54:55-78. doi: 10.1146/annurev-phyto-080615-100217. Epub 2016 May 25. Annu Rev Phytopathol. 2016. PMID: 27296148 Review. - Mechanisms of tomato mosaic virus RNA replication and its inhibition by the host resistance factor Tm-1.
Ishibashi K, Ishikawa M. Ishibashi K, et al. Curr Opin Virol. 2014 Dec;9:8-13. doi: 10.1016/j.coviro.2014.08.005. Epub 2014 Sep 17. Curr Opin Virol. 2014. PMID: 25212767 Review.
Cited by
- A vicilin-like seed storage protein, PAP85, is involved in tobacco mosaic virus replication.
Chen CE, Yeh KC, Wu SH, Wang HI, Yeh HH. Chen CE, et al. J Virol. 2013 Jun;87(12):6888-900. doi: 10.1128/JVI.00268-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576511 Free PMC article. - Susceptibility Genes to Plant Viruses.
Garcia-Ruiz H. Garcia-Ruiz H. Viruses. 2018 Sep 10;10(9):484. doi: 10.3390/v10090484. Viruses. 2018. PMID: 30201857 Free PMC article. Review. - Tobacco Mosaic Virus Movement: From Capsid Disassembly to Transport Through Plasmodesmata.
Ibrahim A, Sasaki N, Schoelz JE, Nelson RS. Ibrahim A, et al. Viruses. 2025 Jan 31;17(2):214. doi: 10.3390/v17020214. Viruses. 2025. PMID: 40006969 Free PMC article. Review. - Defining the proximal interaction networks of Arf GTPases reveals a mechanism for the regulation of PLD1 and PI4KB.
Li FL, Wu Z, Gao YQ, Bowling FZ, Franklin JM, Hu C, Suhandynata RT, Frohman MA, Airola MV, Zhou H, Guan KL. Li FL, et al. EMBO J. 2022 Sep 1;41(17):e110698. doi: 10.15252/embj.2022110698. Epub 2022 Jul 17. EMBO J. 2022. PMID: 35844135 Free PMC article. - Genome-wide analysis of tobacco NtTOM1/TOM3 gene family and identification of NtTOM1a/ NtTOM3a response to tobacco mosaic virus.
Wang X, Bai Y, Shen Z, Zhang X, Cai C, Qiao C, Jiang C, Cheng L, Liu D, Yang A. Wang X, et al. BMC Plant Biol. 2024 Oct 10;24(1):942. doi: 10.1186/s12870-024-05632-1. BMC Plant Biol. 2024. PMID: 39385089 Free PMC article.
References
- Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, et al. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials