HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency - PubMed (original) (raw)
. 2011 Dec;7(12):e1002439.
doi: 10.1371/journal.ppat.1002439. Epub 2011 Dec 8.
Karen E Ocwieja, Jane Rasaiyaah, Amanda J Price, Troy L Brady, Shoshannah L Roth, Stéphane Hué, Adam J Fletcher, KyeongEun Lee, Vineet N KewalRamani, Mahdad Noursadeghi, Richard G Jenner, Leo C James, Frederic D Bushman, Greg J Towers
Affiliations
- PMID: 22174692
- PMCID: PMC3234246
- DOI: 10.1371/journal.ppat.1002439
HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency
Torsten Schaller et al. PLoS Pathog. 2011 Dec.
Abstract
Lentiviruses such as HIV-1 traverse nuclear pore complexes (NPC) and infect terminally differentiated non-dividing cells, but how they do this is unclear. The cytoplasmic NPC protein Nup358/RanBP2 was identified as an HIV-1 co-factor in previous studies. Here we report that HIV-1 capsid (CA) binds directly to the cyclophilin domain of Nup358/RanBP2. Fusion of the Nup358/RanBP2 cyclophilin (Cyp) domain to the tripartite motif of TRIM5 created a novel inhibitor of HIV-1 replication, consistent with an interaction in vivo. In contrast to CypA binding to HIV-1 CA, Nup358 binding is insensitive to inhibition with cyclosporine, allowing contributions from CypA and Nup358 to be distinguished. Inhibition of CypA reduced dependence on Nup358 and the nuclear basket protein Nup153, suggesting that CypA regulates the choice of the nuclear import machinery that is engaged by the virus. HIV-1 cyclophilin-binding mutants CA G89V and P90A favored integration in genomic regions with a higher density of transcription units and associated features than wild type virus. Integration preference of wild type virus in the presence of cyclosporine was similarly altered to regions of higher transcription density. In contrast, HIV-1 CA alterations in another patch on the capsid surface that render the virus less sensitive to Nup358 or TRN-SR2 depletion (CA N74D, N57A) resulted in integration in genomic regions sparse in transcription units. Both groups of CA mutants are impaired in replication in HeLa cells and human monocyte derived macrophages. Our findings link HIV-1 engagement of cyclophilins with both integration targeting and replication efficiency and provide insight into the conservation of viral cyclophilin recruitment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Binding of CANTD to Nup358 and effects on HIV-1 infectivity.
A, VSV-G pseudotyped GFP encoding vectors derived from HIV-1 NL4.3 or MVP5180, SIVmac, or MLV were titered on HeLa cells expressing scrambled control (SC) or Nup358 or TRN-SR2 specific shRNA (mean and SD, n = 3). Relative changes from titer on control cells are shown above the bars. B, Western blots detecting Nup358, TRN-SR2 or β-Actin as loading control. C, Schematic representation of HIV-1/SIVmac chimeras and titration of HIV-1 GFP vector bearing an SIVmac CA (HIV-1 SIVCA) or SIVmac bearing HIV-1 CA-p2 (SIV(HIV-1CAp2)) measured on SC, Nup358, or TRN-SR2 depleted HeLa cells (mean and SD, n = 3). D, Measurement of HIV-1 late reverse transcription product (LRT) or 2-LTR circles at indicated time points post infection (p.i.) in HeLa control cells (SC) or cells depleted for Nup358 or TRN-SR2 (mean and SD, n = 3). A parallel sample was used to determine the number of infected cells by flow cytometry 48 h post infection. E, HeLa control cells (SC) or cells depleted for Nup358 or TRN-SR2 were transduced with an MLV vector expressing human CD4 and the neomycin resistance gene and drug selected cell population were infected with NL4.3GFP-IRES. Percentage of infected cells was enumerated at indicated time points by measuring GFP expression using flow cytometry. F, Isothermal titration calorimetry of cyclophilin A (CypA) or Nup358Cyp against the CANTD domains of NL4.3 HIV-1 or SIVmac in the presence or absence of 10 µM cyclosporine (Cs). G, HIV-1 GFP vector titer on CRFK cells expressing empty vector (EV), owl monkey TRIM5 RBCC fused to human CypA (TRIMCypA) or human Nup358Cyp (TRIMNup358) in the presence or absence of 5 µM Cs (mean and SD, n = 3). Protein levels measured by western blot detecting the HA tag with β-Actin as a loading control.
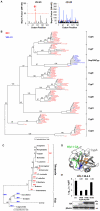
Figure 2. Nup358Cyp evolved under positive selection pressure.
A, Codon positions under positive (dN>dS) and negative (dN<dS) selection along the Nup358Cyp gene sequence, as indicated by the Bayesian probability that a particular site is under selective pressure. A Bayes factor >50 is considered strong evidence for the favored model. B, Maximum likelihood phylogenetic tree of cyclophilins A, B, C, D, E, F, G, H and Nup358Cyp from Rhesus macaque (M. mulatta), Human (H. sapiens), Chimpanzee (P. troglodytes), Dog (C. familiaris), Cow (B. taurus), Horse (E. caballus), Marmoset (C. jacchus), Mouse (M. musculus), Rat (R. norvegicus), Opossum (M. domestica), Zebrafish (D. rerio), Orangutan (P. abelii) and Rabbit (O. cuniculus). Branch lengths and bootstrap supports are shown. C, Maximum likelihood phylogenetic tree demonstrating the simplest model of evolution of residue 61 in Nup358Cyp sequences. D, Structure of CypA bound to CANTD with CypA key residues R55 and M61 as well as HIV-1 CA P90 highlighted. (PDB:1FGL) E, NL4.3 HIV-1 GFP vector titer on CRFK cells expressing empty vector (EV), TRIMCypA, wild type TRIMNup358, or TRIMNup358 mutant V61M. Protein levels measured by western blot detecting the HA tag with β-Actin as loading control are shown. The data are representative of three independent experiments (mean and SD, n = 3).
Figure 3. Effect of CA mutants on Nup358/TRN-SR2 RNAi sensitivity and Nup358Cyp binding.
A, Titers of VSV-G pseudotyped NL4.3 HIV-1 GFP vectors bearing wild type or mutant CA on HeLa control cells (SC), or Nup358 or TRN-SR2 depleted cells in the presence or absence of 2 µg/ml aphidicolin (AC). MLV GFP vector was used as a control for aphidicolin. Relative changes from titer on control cells are shown above the bars. The data are representative of two independent experiments each using three different virus doses. B, ITC of human CypA and Nup358-Cyp against HIV-1 CANTD mutants. Kd values as well as stoichiometries (N), enthalpies (ΔH) and entropies (ΔS) are shown.
Figure 4. Residue changes in HIV-1 CA mutants alter integration site targeting.
A, Effects of CA mutants on integration frequency near multiple chromosomal features. The rows show genomic features, and the columns indicate integration site data sets. The color code indicates ROC areas . The arrow denotes the wild type control set used for pairwise statistical comparisons to the other data sets. P values summarizing the significance of departures from the control are represented by asterisks (*P<0.05; **P<0.01; ***P<0.001). Several different intervals are compared. "Expression density" summarizes the density of genes in the indicated length intervals that are expressed in the upper half ("top 1/2") or upper sixteenth ("top 1/6") of all genes on Affymetrix HU95A chip data for HeLa cells (accessions GSM23372, GSM23373, GSM23377, and GSM23378). For a more detailed guide see . B, ROC area values were used to generate pairwise Euclidean distances, which were then analyzed by hierarchical clustering generating the presented dendogram. The locations of changed residues in mutant viruses studied on C, the CA hexamer (PDB:3GV2) and D, the CA monomer (PDB:3GV2). E, Time course of replication competent NL4.3 (Ba-L Env) bearing wild type, P90A or N74D CA in HeLa TZM-bl cells. Luciferase expression was measured by counting RLU at indicated times after virus inoculation. The data are representative of two independent experiments. F, Replication assay of NL4.3 (Ba-L Env) bearing wild type or CA mutants P90A or N74D in human MDM. Cells were stained for Gag p24 at specific time points after infection and infected colonies counted. Standard errors of the mean of three fields are shown.
Figure 5. CypA inhibition alters HIV-1 integration site targeting.
A, Effects of CypA inhibition by cyclosporine (Cs) on integration frequency near multiple chromosomal features for wild type NL4.3 HIV-1 GFP vector and CA mutants. The arrow denotes the wild type control set used for pairwise statistical comparisons to the other data sets. The heat map analysis was performed as in Figure 4A. B, ROC area values were used to generate pairwise Euclidean distances, which were then analyzed by hierarchical clustering generating the presented dendogram.
Figure 6. CypA inhibition forces HIV-1 into a Nup358/Nup153 independent nuclear entry pathway.
A, Titers of VSV-G pseudotyped NL4.3 HIV-1 GFP vectors bearing wild type or mutant CA on HeLa control cells (SC), or Nup358 or TRN-SR2 depleted cells in the presence or absence of 8 µM Cs (mean and SD, n = 3). Relative changes from titer on control cells are shown above the bars. B, HeLa cells expressing scrambled control (SC), or Nup358 specific shRNA were transiently transduced with either MLV empty vector control or vector expressing CypA specific shRNA and subsequently infected with wild type NL4.3 HIV-1 GFP vector or CA mutant P90A or O-group MVP5180 GFP vector. Infection was measured 48 hours later by FACS to enumerate infected cells. Fold changes to control cells are shown. A parallel western blot for experiments in B detecting CypA and β-Actin as loading control. C, HeLa control cells (SC) or transiently Nup153 depleted cells were infected with indicated vectors in the presence or absence of 8 µM Cs (Mean and SD, n = 3). Western blot detecting Nup153 and β-Actin as a loading control. D, MDM from 2 independent donors were infected with 400 pg RT replication competent HIV-1 NL4.3 (Ba-L Env) in the presence of 5 µM Cs or DMSO. Replication was measured over time by counting CA positive cells after staining with a CA specific antibody.
Figure 7. Model for CypA-dependent/-independent HIV-1 nuclear entry pathways.
A, Wild type (WT) HIV-1 capsids are bound by cytoplasmic cyclophilin A (CypA) molecules that direct the capsid to use a specific nuclear entry pathway. The nuclear entry pathway involves CA binding to the cyclophilin domain of the cytoplasmic nuclear pore complex (NPC) component Nup358. Nup358 may mediate uncoating directly at the nuclear pore, liberating the pre-integration complex, which will interact with TRN-SR2 and the nuclear NPC component Nup153. B, Inhibition of CypA-CA interaction by the drug cyclosporine (Cs) or by substitution of CA residues that abolish CypA binding directs the virus into a less productive CypA-independent nuclear entry pathway that displays impaired dependence on Nup358 and Nup153. This route of nuclear entry results in virus integration preferences in areas of higher density of transcription units and associated features. C, HIV-1 capsid mutants that are less sensitive to Nup358, Nup153 or TRN-SR2 RNAi (HIV-1 CA N57A or N74D) enter the nucleus through a different pathway that directs their integration into genome areas of lower density of transcription units. D, Depletion of Nup358 by RNAi reduces viral nuclear entry via the Nup358-dependent pathway and the virus gains access via an Nup358-independent alternative pathway resulting in phenotypically similar integration site selection as observed for the CA mutants N74D or N57A. Alternative nuclear entry pathways disturb HIV-1 integration site selection, possibly contributing to sub-optimal replication of the virus in spreading infection assays.
Similar articles
- KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection.
Dharan A, Talley S, Tripathi A, Mamede JI, Majetschak M, Hope TJ, Campbell EM. Dharan A, et al. PLoS Pathog. 2016 Jun 21;12(6):e1005700. doi: 10.1371/journal.ppat.1005700. eCollection 2016 Jun. PLoS Pathog. 2016. PMID: 27327622 Free PMC article. - A cyclophilin homology domain-independent role for Nup358 in HIV-1 infection.
Meehan AM, Saenz DT, Guevera R, Morrison JH, Peretz M, Fadel HJ, Hamada M, van Deursen J, Poeschla EM. Meehan AM, et al. PLoS Pathog. 2014 Feb 20;10(2):e1003969. doi: 10.1371/journal.ppat.1003969. eCollection 2014 Feb. PLoS Pathog. 2014. PMID: 24586169 Free PMC article. - MxB impedes the NUP358-mediated HIV-1 pre-integration complex nuclear import and viral replication cooperatively with CPSF6.
Xie L, Chen L, Zhong C, Yu T, Ju Z, Wang M, Xiong H, Zeng Y, Wang J, Hu H, Hou W, Feng Y. Xie L, et al. Retrovirology. 2020 Jun 29;17(1):16. doi: 10.1186/s12977-020-00524-2. Retrovirology. 2020. PMID: 32600399 Free PMC article. - A model for cofactor use during HIV-1 reverse transcription and nuclear entry.
Hilditch L, Towers GJ. Hilditch L, et al. Curr Opin Virol. 2014 Feb;4(100):32-6. doi: 10.1016/j.coviro.2013.11.003. Epub 2014 Jan 14. Curr Opin Virol. 2014. PMID: 24525292 Free PMC article. Review. - HIV Capsid and Integration Targeting.
Engelman AN. Engelman AN. Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review.
Cited by
- Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication.
Di Nunzio F, Fricke T, Miccio A, Valle-Casuso JC, Perez P, Souque P, Rizzi E, Severgnini M, Mavilio F, Charneau P, Diaz-Griffero F. Di Nunzio F, et al. Virology. 2013 May 25;440(1):8-18. doi: 10.1016/j.virol.2013.02.008. Epub 2013 Mar 21. Virology. 2013. PMID: 23523133 Free PMC article. - CPSF6 defines a conserved capsid interface that modulates HIV-1 replication.
Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, KewalRamani VN, Chin JW, Towers GJ, James LC. Price AJ, et al. PLoS Pathog. 2012;8(8):e1002896. doi: 10.1371/journal.ppat.1002896. Epub 2012 Aug 30. PLoS Pathog. 2012. PMID: 22956906 Free PMC article. - Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells.
Hulme AE, Kelley Z, Foley D, Hope TJ. Hulme AE, et al. J Virol. 2015 May;89(10):5350-61. doi: 10.1128/JVI.00476-15. Epub 2015 Mar 4. J Virol. 2015. PMID: 25741002 Free PMC article. - Targeting the cyclophilin domain of Ran-binding protein 2 (Ranbp2) with novel small molecules to control the proteostasis of STAT3, hnRNPA2B1 and M-opsin.
Cho KI, Orry A, Park SE, Ferreira PA. Cho KI, et al. ACS Chem Neurosci. 2015 Aug 19;6(8):1476-85. doi: 10.1021/acschemneuro.5b00134. Epub 2015 Jun 12. ACS Chem Neurosci. 2015. PMID: 26030368 Free PMC article. - HIV Genome-Wide Protein Associations: a Review of 30 Years of Research.
Li G, De Clercq E. Li G, et al. Microbiol Mol Biol Rev. 2016 Jun 29;80(3):679-731. doi: 10.1128/MMBR.00065-15. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27357278 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- G0600081/MRC_/Medical Research Council/United Kingdom
- G9721629/MRC_/Medical Research Council/United Kingdom
- R01 AI082020/AI/NIAID NIH HHS/United States
- AI52845/AI/NIAID NIH HHS/United States
- R01 AI052845/AI/NIAID NIH HHS/United States
- AI082020/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- G0900950/MRC_/Medical Research Council/United Kingdom
- MC_U105181010/MRC_/Medical Research Council/United Kingdom
- 090940/WT_/Wellcome Trust/United Kingdom
- G0801172/MRC_/Medical Research Council/United Kingdom
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous