HumMeth27QCReport: an R package for quality control and primary analysis of Illumina Infinium methylation data - PubMed (original) (raw)
HumMeth27QCReport: an R package for quality control and primary analysis of Illumina Infinium methylation data
Francesco M Mancuso et al. BMC Res Notes. 2011.
Abstract
Background: The study of the human DNA methylome has gained particular interest in the last few years. Researchers can nowadays investigate the potential role of DNA methylation in common disorders by taking advantage of new high-throughput technologies. Among these, Illumina Infinium assays can interrogate the methylation levels of hundreds of thousands of CpG sites, offering an ideal solution for genome-wide methylation profiling. However, like for other high-throughput technologies, the main bottleneck remains at the stage of data analysis rather than data production.
Findings: We have developed HumMeth27QCReport, an R package devoted to researchers wanting to quickly analyse their Illumina Infinium methylation arrays. This package automates quality control steps by generating a report including sample-independent and sample-dependent quality plots, and performs primary analysis of raw methylation calls by computing data normalization, statistics, and sample similarities. This package is available at CRAN repository, and can be integrated in any Galaxy instance through the implementation of ad-hoc scripts accessible at Galaxy Tool Shed.
Conclusions: Our package provides users of the Illumina Infinium Methylation assays with a simplified, automated, open-source quality control and primary analysis of their methylation data. Moreover, to enhance its use by experimental researchers, the tool is being distributed along with the scripts necessary for its implementation in the Galaxy workbench. Finally, although it was originally developed for HumanMethylation27, we proved its compatibility with data generated with the HumanMethylation450 Bead Chip.
Figures
Figure 1
HumMeth27QCReport pipeline. Standard analysis workflow of Illumina Methylation Assay data. Steps facilitated by H_umMeth27QCReport_ are included in dark gray box.
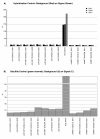
Figure 2
Internal controls. Examples of sample-independent (A) and sample-dependent (B) plots generated by the HumMeth27QCReport package. a. Hybridization control, to check synthetic targets at low, medium, and high concentration level. Low percentage for each level is an indication of good performance; b. bisulfite control on green channel, to monitor the efficiency of genomic-DNA bisulfite conversion using the Infinium I assay design. Low percentage is an indication of good performance, whereas if the percentage is high we consider that the sample has lower performance. A detailed explanation of each plot is available at the HumMeth27QCReport homepage (
http://biocore.crg.es/wiki/HumMeth27QCReport
). JUR: Jurkat DNA; JUR MET: methylated Jurkat DNA; UNMET: unmethylated DNA; EZGOLD: EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA); QIAGEN: EpiTect Bisulfite Kit (QiaGen); BIO1: biological replicate 1; BIO2: biological replicate 2; PD3 Amyg: example of sample of poor quality.
Similar articles
- Ewastools: Infinium Human Methylation BeadChip pipeline for population epigenetics integrated into Galaxy.
Murat K, Grüning B, Poterlowicz PW, Westgate G, Tobin DJ, Poterlowicz K. Murat K, et al. Gigascience. 2020 May 1;9(5):giaa049. doi: 10.1093/gigascience/giaa049. Gigascience. 2020. PMID: 32401319 Free PMC article. - Marmal-aid--a database for Infinium HumanMethylation450.
Lowe R, Rakyan VK. Lowe R, et al. BMC Bioinformatics. 2013 Dec 12;14:359. doi: 10.1186/1471-2105-14-359. BMC Bioinformatics. 2013. PMID: 24330312 Free PMC article. - Global DNA methylation profiling technologies and the ovarian cancer methylome.
Tang J, Fang F, Miller DF, Pilrose JM, Matei D, Huang TH, Nephew KP. Tang J, et al. Methods Mol Biol. 2015;1238:653-75. doi: 10.1007/978-1-4939-1804-1_34. Methods Mol Biol. 2015. PMID: 25421685 Review. - Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis.
Sun Z, Cunningham J, Slager S, Kocher JP. Sun Z, et al. Epigenomics. 2015 Aug;7(5):813-28. doi: 10.2217/epi.15.21. Epub 2015 Sep 14. Epigenomics. 2015. PMID: 26366945 Free PMC article. Review.
Cited by
- Identification of DNA methylation biomarkers from Infinium arrays.
Wessely F, Emes RD. Wessely F, et al. Front Genet. 2012 Aug 25;3:161. doi: 10.3389/fgene.2012.00161. eCollection 2012. Front Genet. 2012. PMID: 22936948 Free PMC article. - Nonpromoter methylation of the CDKN2A gene with active transcription is associated with improved locoregional control in laryngeal squamous cell carcinoma.
Ben-Dayan MM, Ow TJ, Belbin TJ, Wetzler J, Smith RV, Childs G, Diergaarde B, Hayes DN, Grandis JR, Prystowsky MB, Schlecht NF. Ben-Dayan MM, et al. Cancer Med. 2017 Feb;6(2):397-407. doi: 10.1002/cam4.961. Epub 2017 Jan 19. Cancer Med. 2017. PMID: 28102032 Free PMC article. - Review of processing and analysis methods for DNA methylation array data.
Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, Marsit CJ, Houseman EA, Brown R. Wilhelm-Benartzi CS, et al. Br J Cancer. 2013 Sep 17;109(6):1394-402. doi: 10.1038/bjc.2013.496. Epub 2013 Aug 27. Br J Cancer. 2013. PMID: 23982603 Free PMC article. Review. - Performance in omics analyses of blood samples in long-term storage: opportunities for the exploitation of existing biobanks in environmental health research.
Hebels DG, Georgiadis P, Keun HC, Athersuch TJ, Vineis P, Vermeulen R, Portengen L, Bergdahl IA, Hallmans G, Palli D, Bendinelli B, Krogh V, Tumino R, Sacerdote C, Panico S, Kleinjans JC, de Kok TM, Smith MT, Kyrtopoulos SA; EnviroGenomarkers Project Consortium. Hebels DG, et al. Environ Health Perspect. 2013 Apr;121(4):480-7. doi: 10.1289/ehp.1205657. Epub 2013 Feb 5. Environ Health Perspect. 2013. PMID: 23384616 Free PMC article. - BIFI: a Taverna plugin for a simplified and user-friendly workflow platform.
Yildiz A, Dilaveroglu E, Visne I, Günay B, Sefer E, Weinhausel A, Rattay F, Goble CA, Pandey RV, Kriegner A. Yildiz A, et al. BMC Res Notes. 2014 Oct 20;7:740. doi: 10.1186/1756-0500-7-740. BMC Res Notes. 2014. PMID: 25332013 Free PMC article.
References
LinkOut - more resources
Full Text Sources