The dependence of viral RNA replication on co-opted host factors - PubMed (original) (raw)
Review
The dependence of viral RNA replication on co-opted host factors
Peter D Nagy et al. Nat Rev Microbiol. 2011.
Abstract
Positive-sense RNA ((+)RNA) viruses such as hepatitis C virus exploit host cells by subverting host proteins, remodelling subcellular membranes, co-opting and modulating protein and ribonucleoprotein complexes, and altering cellular metabolic pathways during infection. To facilitate RNA replication, (+)RNA viruses interact with numerous host molecules through protein-protein, RNA-protein and protein-lipid interactions. These interactions lead to the formation of viral replication complexes, which produce new viral RNA progeny in host cells. This Review presents the recent progress that has been made in understanding the role of co-opted host proteins and membranes during (+)RNA virus replication, and discusses common themes employed by different viruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
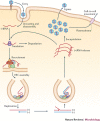
Figure 1. Schematic infection cycle of positive-sense RNA viruses.
Positive-sense RNA ((+)RNA) viruses enter animal cells by endocytosis and plant cells through wounds. When the virus is inside the cell, the (+)RNA genome is released into the cytosol, where it is translated by the host ribosomes. The resulting viral replication proteins then recruit the (+)RNA to subcellular membrane compartments, where functional viral replication complexes (VRCs) are assembled. A small amount of negative-sense RNA ((−)RNA) is synthesized and serves as a template for the synthesis of a large number of (+)RNA progeny. The new (+)RNAs are released from the VRCs, whereas the (−)RNA is retained. The released (+)RNAs start a new cycle of translation and replication, become encapsidated, and then exit the cells (in the case of animal viruses) or move to neighbouring cells through plasmodesmata (in the case of plant viruses).
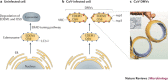
Figure 2. Remodelling the cellular secretory pathway to support human poliovirus replication.
a | The human poliovirus (PV) viral replication complex (VRC) assembles on the Golgi–_trans_-Golgi network (TGN) membrane and contains proteolytically cleaved host poly(rC)-binding protein 2 (PCBP2) and the virally encoded RNA-dependent RNA polymerase (RdRp) precursor protein, 3CD, bound to the 5′ cloverleaf-like structure of the positive-sense RNA ((+)RNA), with a host poly(A)-binding protein (PABP) bound to the 3′ poly(A) tail. As a result of the interaction between PCBP2 and PABP, the ends of the PV RNA genome are brought into proximity, facilitating the cleavage of 3CD and the release of the RdRp (3D-POL). 3D-POL then starts synthesis of the negative-sense RNA ((–)RNA) using VPg–UU (di-uridylated VPg protein) as a primer. The newly made double-stranded RNA (dsRNA) replicative form (RF), which consists of hybridized (+)RNA and (−)RNA, is partly unwound by the chaperone activity of the host heterogeneous nuclear ribonucleoprotein C (hnRNPC) and the binding of PCBP2 and 3CD to the 5′ end of the (+)RNA. After cleavage of 3CD, the released 3D-POL starts (+)RNA synthesis, which will go on for many rounds to produce excess amounts of (+)RNA progeny. b | PV begins replicating in the Golgi–TGN compartment. The virally encoded tail-anchored membrane protein 3A binds to the small GTPase ADP-ribosylation factor 1 (ARF1) and promotes the degradation of the host general vesicular transport protein p115, preventing the recruitment of ɛ-COP and β-COP (components of the vesicle coatomer complex 1, which surrounds transport vesicles in the early secretory pathway) to ERGIC. These events slow down vesicular transport from the endoplasmic reticulum (ER) to the Golgi–TGN. The production of additional 3A molecules on the newly made PV (+)RNAs results in remodelling of the Golgi–TGN and ERGIC compartments into viral replication organelles (the VRCs). These organelles provide optimal replication for PV owing to the presence of an increased amount of the lipid phosphatidylinositol 4-phosphate (PtdIns4P), which is produced by the recruited host PtdIns4P synthesis enzyme, PtdIns4-kinase-β (PI4KIIIβ). c | Trafficking between the ER and the Golgi in uninfected cells. The ERGIC is decorated with COP1, which is recruited by ARF1 and its guanine nucleotide exchange factor, GBF1.
Figure 3. Structures of the flavivirus replication complexes.
a | Hepatitis C virus (HCV) replication takes place at a unique subcellular compartment, the endoplasmic reticulum (ER)-derived membranous web, which consists of clusters of HCV-induced vesicles and lipid droplets (LD) and is the proposed site of viral assembly. The virus-encoded replication protein non-structural protein 5A (NS5A) binds to human vesicle-associated membrane protein-associated protein A (VAPA), which, together with NS4B, seems to be important for the association of NS5A and NS5B with cholesterol-rich lipid rafts in the ER-derived membranes. The membrane-bound NS5A then recruits and activates the host kinase phosphatidylinositol 4-kinase-α (PI4KIIIα), which leads to an enrichment of phosphatidylinositol-4-phosphate (PtdIns4P) that could facilitate recruitment of oxysterol-binding protein 1 (OSBP1) and the formation of the sterol-rich 'membranous web' to support HCV replication. The high concentration of PtdIns4P probably also facilitates the recruitment of additional host factors to the membranous web. The viral protein NS4B is also important for the formation of the membranous web, possibly by inducing membrane curvature (not shown). b | Electron micrograph of the viral replication complexes (VRCs) induced during dengue virus (DENV) infections. The white arrowhead indicates a DENV-induced vesicle in the ER with an opening towards the nuclear envelope; the black arrowhead points to a virus particle in a cisterna close to a Golgi stack. c | Three-dimensional modelling of the VRCs induced during DENV infections. The arrow indicates a putative virus budding site, and red spheres represent virus particles. ARF1, ADP-ribosylation factor 1; NE, nuclear envelope; Ve, vesicle. Parts b,c are reproduced, with permission, from Ref. © (2009) Elsevier.
Figure 4. Proposed remodelling of the endoplasmic reticulum-associated protein degradation tuning pathway during coronavirus replication.
a | In normal, uninfected cells, the endoplasmic reticulum-associated protein degradation (ERAD) tuning pathway downregulates the levels of the ER chaperones EDEM1 (ER degradation-enhancing α-mannosidase-like 1) and OS9 in the ER via transport to the lysosome, mediated by LC3-I (non-lipidated microtubule-associated proteins 1A and 1B light chain 3A)-coated edemosomes. b | Coronavirus (CoV) infection transforms the edemosomes into double-membraned vesicles (DMVs) that probably contain viral replication complexes (VRCs). The actual orientation of the CoV VRCs in DMVs is not yet known. c | Electron tomography-based model of CoV DMVs. Narrow connections between several DMVs form a network that is also connected to the ER. Part c is reproduced from Ref. .
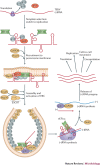
Figure 5. Functions of host factors during tombusvirus replication in Saccharomyces cerevisiae.
Tombusvirus (such as tomato bushy stunt virus (TBSV)) RNA translation and replication, and the host proteins that affect these processes in a Saccharomyces cerevisiae model host. The TBSV RNA-dependent RNA polymerase (RdRp) precursor p92 is activated during the assembly process (as indicated by the colour change) in an as-yet-unknown way. TBSV p33 is an RNA chaperone involved in viral RNA recruitment and assembly of the viral replication complexes (VRCs). The presence of host proteins (denoted host factor (HF) here) is currently hypothetical, and they could be ESCRT-III (endosomal sorting complexes required for transport III) factors. The known cellular functions for the shown host proteins are: elongation factor 1α (eEF1α), required for translation elongation; Pex19, a cytosolic shuttle protein involved in transporting peroxisomal membrane proteins to the peroxisome; heat shock protein 70 (Hsp70), a molecular chaperone involved in protein folding, mostly in the cytosol; Erg25, an enzyme for sterol biosynthesis; Cpr1 (also called cyclophilin), a cytosolic prolyl isomerase; ESCRT, a large family of proteins involved in membrane bending, protein transport and degradation via the endosomal pathway; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzymes, a ubiquitous protein family that is a key component of cytosolic energy production; Ded1, a DEAD box helicase involved in translation initiation.
Similar articles
- Co-opting the fermentation pathway for tombusvirus replication: Compartmentalization of cellular metabolic pathways for rapid ATP generation.
Lin W, Liu Y, Molho M, Zhang S, Wang L, Xie L, Nagy PD. Lin W, et al. PLoS Pathog. 2019 Oct 24;15(10):e1008092. doi: 10.1371/journal.ppat.1008092. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31648290 Free PMC article. - Host factors in the replication of positive-strand RNA viruses.
Wang RY, Li K. Wang RY, et al. Chang Gung Med J. 2012 Mar-Apr;35(2):111-24. doi: 10.4103/2319-4170.106160. Chang Gung Med J. 2012. PMID: 22537926 Review. - MicroRNA Regulation of RNA Virus Replication and Pathogenesis.
Trobaugh DW, Klimstra WB. Trobaugh DW, et al. Trends Mol Med. 2017 Jan;23(1):80-93. doi: 10.1016/j.molmed.2016.11.003. Epub 2016 Dec 16. Trends Mol Med. 2017. PMID: 27989642 Free PMC article. Review. - Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.
Lloyd RE. Lloyd RE. Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review. - Diverse roles of host RNA binding proteins in RNA virus replication.
Li Z, Nagy PD. Li Z, et al. RNA Biol. 2011 Mar-Apr;8(2):305-15. doi: 10.4161/rna.8.2.15391. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21505273 Free PMC article. Review.
Cited by
- Phosphatidic acid produced by phospholipase D promotes RNA replication of a plant RNA virus.
Hyodo K, Taniguchi T, Manabe Y, Kaido M, Mise K, Sugawara T, Taniguchi H, Okuno T. Hyodo K, et al. PLoS Pathog. 2015 May 28;11(5):e1004909. doi: 10.1371/journal.ppat.1004909. eCollection 2015 May. PLoS Pathog. 2015. PMID: 26020241 Free PMC article. - Viral sensing of the subcellular environment regulates the assembly of new viral replicase complexes during the course of infection.
Nagy PD. Nagy PD. J Virol. 2015 May;89(10):5196-9. doi: 10.1128/JVI.02973-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25741009 Free PMC article. Review. - Co-opting the fermentation pathway for tombusvirus replication: Compartmentalization of cellular metabolic pathways for rapid ATP generation.
Lin W, Liu Y, Molho M, Zhang S, Wang L, Xie L, Nagy PD. Lin W, et al. PLoS Pathog. 2019 Oct 24;15(10):e1008092. doi: 10.1371/journal.ppat.1008092. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31648290 Free PMC article. - What proteomics can reveal about plant-virus interactions? Photosynthesis-related proteins on the spotlight.
Souza PFN, Garcia-Ruiz H, Carvalho FEL. Souza PFN, et al. Theor Exp Plant Physiol. 2019 Mar;31(1):227-248. doi: 10.1007/s40626-019-00142-0. Epub 2019 Jan 22. Theor Exp Plant Physiol. 2019. PMID: 31355128 Free PMC article. - Sugarcane mosaic virus orchestrates the lactate fermentation pathway to support its successful infection.
Jiang T, Du K, Wang P, Wang X, Zang L, Peng D, Chen X, Sun G, Zhang H, Fan Z, Cao Z, Zhou T. Jiang T, et al. Front Plant Sci. 2023 Jan 9;13:1099362. doi: 10.3389/fpls.2022.1099362. eCollection 2022. Front Plant Sci. 2023. PMID: 36699858 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases