Nature of curvature coupling of amphiphysin with membranes depends on its bound density - PubMed (original) (raw)
Nature of curvature coupling of amphiphysin with membranes depends on its bound density
Benoît Sorre et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Cells are populated by a vast array of membrane-binding proteins that execute critical functions. Functions, like signaling and intracellular transport, require the abilities to bind to highly curved membranes and to trigger membrane deformation. Among these proteins is amphiphysin 1, implicated in clathrin-mediated endocytosis. It contains a Bin-Amphiphysin-Rvs membrane-binding domain with an N-terminal amphipathic helix that senses and generates membrane curvature. However, an understanding of the parameters distinguishing these two functions is missing. By pulling a highly curved nanotube of controlled radius from a giant vesicle in a solution containing amphiphysin, we observed that the action of the protein depends directly on its density on the membrane. At low densities of protein on the nearly flat vesicle, the distribution of proteins and the mechanical effects induced are described by a model based on spontaneous curvature induction. The tube radius and force are modified by protein binding but still depend on membrane tension. In the dilute limit, when practically no proteins were present on the vesicle, no mechanical effects were detected, but strong protein enrichment proportional to curvature was seen on the tube. At high densities, the radius is independent of tension and vesicle protein density, resulting from the formation of a scaffold around the tube. As a consequence, the scaling of the force with tension is modified. For the entire density range, protein was enriched on the tube as compared to the vesicle. Our approach shows that the strength of curvature sensing and mechanical effects on the tube depends on the protein density.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
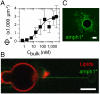
Fig. 1.
Two regimes for amphiphysin 1 binding to a GUV membrane DOPC∶DOPE∶DOPS (1∶1∶1). (A) Adsorption isotherm of amphiphysin 1 on GUV. The amphiphysin density, Φ_ν_, deduced from the fluorescence signal, as a function of the protein bulk density C_bulk. Data are fitted with Φ_v = Φmax/(1 + _K_d/_C_bulk) with Φmax = 3,000 μm-2 and _K_d = 35 nm (error bars correspond to standard deviation. N = 6 vesicles for each point). (B) For _C_bulk < _K_d, the amphiphysin signal (green) is undetectable on the GUV, the fluorescent lipid signal is in red, and amphiphysin is visible on the tube. (C) At high _C_bulk (here equal to 1 μM) amphiphysin binds to the GUV and forms tubes rich in amphiphysin (green fluorescence). (Scale bar: 5 μm.)
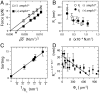
Fig. 2.
Low-density regime. (A) Force curve shift due to amphiphysin binding in the low-density regime. Here, Φ_v_ = 280 ± 100 μm-2. The force is lower with protein (▪) than without (□). Data are fitted to  , where A = 55 ± 2 pN1/2·nm1/2 and B = 0 without protein and A = 52 ± 2 × pN1/2·nm1/2 and B = -3.5 ± 2 pN with protein. (B) The radius, R t, versus tension, σ, with protein (○) or without (●). Radius is deduced from fluorescence. (C) Linear variation of the sorting ratio as a function of 1/R t. Data correspond to five independent experiments. A fit using Eq. 3 gives
, where A = 55 ± 2 pN1/2·nm1/2 and B = 0 without protein and A = 52 ± 2 × pN1/2·nm1/2 and B = -3.5 ± 2 pN with protein. (B) The radius, R t, versus tension, σ, with protein (○) or without (●). Radius is deduced from fluorescence. (C) Linear variation of the sorting ratio as a function of 1/R t. Data correspond to five independent experiments. A fit using Eq. 3 gives  . (D) Variation of R t, at fixed tension σ = 2 × 10-5 N/m, versus Φ. The data are fitted to R t = 32 - (38 ± 5) × 10-3Φ_v_ nm (line), as deduced from Eq. 2. Round symbol corresponds to expected radius for a 10 k_B_T membrane (32 nm).
. (D) Variation of R t, at fixed tension σ = 2 × 10-5 N/m, versus Φ. The data are fitted to R t = 32 - (38 ± 5) × 10-3Φ_v_ nm (line), as deduced from Eq. 2. Round symbol corresponds to expected radius for a 10 k_B_T membrane (32 nm).
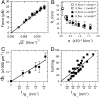
Fig. 3.
The dilute limit. The results presented in this figure correspond to a single GUV with Φ_v_ < 50 μm-2. (A) No difference between the tube force as a function of  with protein (□) or without (▪). The linear fit to the force,
with protein (□) or without (▪). The linear fit to the force,  , gives κ = 12 ± 2 k_B_T. (B) The radius, R t, versus tension, σ, with protein (empty symbols) or without (full symbols). Radius is deduced either from fluorescence (round symbols) or from force (square symbols) measurements. (C) Amphiphysin density on the tube, Φ_t_, versus tube curvature, 1/R t. R t was found from force measurements. A linear fit yields Φ_t_ = A/R t μm-2, where A = 29 ± 2 μm-1. (D) Linear variation of the sorting ratio as a function of 1/R t. Data correspond to five independent experiments. A fit using Eq. 4 gives
, gives κ = 12 ± 2 k_B_T. (B) The radius, R t, versus tension, σ, with protein (empty symbols) or without (full symbols). Radius is deduced either from fluorescence (round symbols) or from force (square symbols) measurements. (C) Amphiphysin density on the tube, Φ_t_, versus tube curvature, 1/R t. R t was found from force measurements. A linear fit yields Φ_t_ = A/R t μm-2, where A = 29 ± 2 μm-1. (D) Linear variation of the sorting ratio as a function of 1/R t. Data correspond to five independent experiments. A fit using Eq. 4 gives  .
.
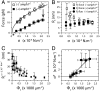
Fig. 4.
High-density regime. The experiments presented in A and B correspond to a single GUV with Φ_v_ = 1,100 ± 100 μm-2. (A) The force is lower with protein (▪) than without (□). Force data without protein are fitted to  , where A = 56 ± 1 pN1/2 nm1/2, and without to f = B(σ - σ_∗), where B = 67 ± 4 nm and σ_∗ = (1.0 ± 0.9) × 10-5 N/m (the asterisk denotes σ_∗). (B) R t versus σ with no protein (empty symbols) and with protein (full symbols). The radius is found from fluorescence (round symbols) or from force (square symbols) measurements. With no protein, the radius was determined from the force according to R t = f/4_πσ; in its presence, the radius was found using R t = f/2_πσ. (C) R t as a function of Φ_v, as measured by fluorescence. (D) Tension at zero force, σ_∗, versus Φ_v. Data were fitted to Eq. 6, neglecting the logarithmic term, _σ_∗ = A_Φ_v + B, where A = (4 ± 1) × 10-8 Nm-1 μm-2 and B = (-2.7 ± 1.4) × 10-5 Nm-1.
, where A = 56 ± 1 pN1/2 nm1/2, and without to f = B(σ - σ_∗), where B = 67 ± 4 nm and σ_∗ = (1.0 ± 0.9) × 10-5 N/m (the asterisk denotes σ_∗). (B) R t versus σ with no protein (empty symbols) and with protein (full symbols). The radius is found from fluorescence (round symbols) or from force (square symbols) measurements. With no protein, the radius was determined from the force according to R t = f/4_πσ; in its presence, the radius was found using R t = f/2_πσ. (C) R t as a function of Φ_v, as measured by fluorescence. (D) Tension at zero force, σ_∗, versus Φ_v. Data were fitted to Eq. 6, neglecting the logarithmic term, _σ_∗ = A_Φ_v + B, where A = (4 ± 1) × 10-8 Nm-1 μm-2 and B = (-2.7 ± 1.4) × 10-5 Nm-1.
Similar articles
- Quantifying Membrane Curvature Generation of Drosophila Amphiphysin N-BAR Domains.
Heinrich MC, Capraro BR, Tian A, Isas JM, Langen R, Baumgart T. Heinrich MC, et al. J Phys Chem Lett. 2010 Nov 16;1(23):3401-3406. doi: 10.1021/jz101403q. J Phys Chem Lett. 2010. PMID: 23772271 Free PMC article. - FBAR syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner.
Ramesh P, Baroji YF, Reihani SN, Stamou D, Oddershede LB, Bendix PM. Ramesh P, et al. Sci Rep. 2013;3:1565. doi: 10.1038/srep01565. Sci Rep. 2013. PMID: 23535634 Free PMC article. - Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2.
Neumann S, Schmid SL. Neumann S, et al. J Biol Chem. 2013 Aug 30;288(35):25119-25128. doi: 10.1074/jbc.M113.490474. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861397 Free PMC article. - When Physics Takes Over: BAR Proteins and Membrane Curvature.
Simunovic M, Voth GA, Callan-Jones A, Bassereau P. Simunovic M, et al. Trends Cell Biol. 2015 Dec;25(12):780-792. doi: 10.1016/j.tcb.2015.09.005. Epub 2015 Oct 28. Trends Cell Biol. 2015. PMID: 26519988 Free PMC article. Review. - Subcellular membrane curvature mediated by the BAR domain superfamily proteins.
Suetsugu S, Toyooka K, Senju Y. Suetsugu S, et al. Semin Cell Dev Biol. 2010 Jun;21(4):340-9. doi: 10.1016/j.semcdb.2009.12.002. Epub 2009 Dec 4. Semin Cell Dev Biol. 2010. PMID: 19963073 Review.
Cited by
- Interferon-Induced Transmembrane Protein 3 Blocks Fusion of Diverse Enveloped Viruses by Altering Mechanical Properties of Cell Membranes.
Guo X, Steinkühler J, Marin M, Li X, Lu W, Dimova R, Melikyan GB. Guo X, et al. ACS Nano. 2021 May 25;15(5):8155-8170. doi: 10.1021/acsnano.0c10567. Epub 2021 Mar 3. ACS Nano. 2021. PMID: 33656312 Free PMC article. - How Membrane Geometry Regulates Protein Sorting Independently of Mean Curvature.
Larsen JB, Rosholm KR, Kennard C, Pedersen SL, Munch HK, Tkach V, Sakon JJ, Bjørnholm T, Weninger KR, Bendix PM, Jensen KJ, Hatzakis NS, Uline MJ, Stamou D. Larsen JB, et al. ACS Cent Sci. 2020 Jul 22;6(7):1159-1168. doi: 10.1021/acscentsci.0c00419. Epub 2020 Jun 23. ACS Cent Sci. 2020. PMID: 32724850 Free PMC article. - Value of models for membrane budding.
Lee CT, Akamatsu M, Rangamani P. Lee CT, et al. Curr Opin Cell Biol. 2021 Aug;71:38-45. doi: 10.1016/j.ceb.2021.01.011. Epub 2021 Mar 8. Curr Opin Cell Biol. 2021. PMID: 33706232 Free PMC article. Review. - Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner.
Tsai FC, Bertin A, Bousquet H, Manzi J, Senju Y, Tsai MC, Picas L, Miserey-Lenkei S, Lappalainen P, Lemichez E, Coudrier E, Bassereau P. Tsai FC, et al. Elife. 2018 Sep 20;7:e37262. doi: 10.7554/eLife.37262. Elife. 2018. PMID: 30234483 Free PMC article. - Guided by curvature: shaping cells by coupling curved membrane proteins and cytoskeletal forces.
Gov NS. Gov NS. Philos Trans R Soc Lond B Biol Sci. 2018 May 26;373(1747):20170115. doi: 10.1098/rstb.2017.0115. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29632267 Free PMC article. Review.
References
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. - PubMed
- Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials