Template-dependent polymerization across discontinuous templates by the heterodimeric primase from the hyperthermophilic archaeon Sulfolobus solfataricus - PubMed (original) (raw)
Template-dependent polymerization across discontinuous templates by the heterodimeric primase from the hyperthermophilic archaeon Sulfolobus solfataricus
Jinchuan Hu et al. Nucleic Acids Res. 2012 Apr.
Abstract
The eukaryotic-like primase from the hyperthermophilic archaeon Sulfolobus solfataricus (SsoPriSL) exhibits a range of activities including template-dependent de novo primer synthesis, primer extension and template-independent terminal nucleotidyl transfer using either rNTPs or dNTPs. Remarkably, the enzyme is able to synthesize products far longer than templates in vitro. Here we show that the long products resulted from template-dependent polymerization across discontinuous templates (PADT) by SsoPriSL. PADT was initiated through either primer synthesis or terminal transfer, and occurred efficiently on templates containing contiguous dCs. Template switching took place when the 3'-end of a growing strand synthesized on one template annealed to another template directly or following the terminal addition of nucleotides, and was subsequently extended on the new template. The key to PADT was the ability of SsoPriSL to promote strand annealing. SsoPriSL catalyzed PADT with either dNTPs or rNTPs as the substrates but preferred the latter. The enzyme remained active in PADT but became inefficient in primer synthesis in vitro when temperature was raised from 55°C to 70°C. Our results suggest that SsoPriSL is capable of bridging noncomplementary DNA ends and, therefore, may serve a role in double-strand DNA break repair in Archaea.
Figures
Figure 1.
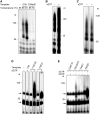
SsoPriSL synthesized products much longer than templates in a template-dependent manner. (A) Comparison of SsoPriSL activity on oligo(dC) and oligo(dT). SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with oligo(dC) (0.25 μM) and [α-32P]rGTP (10 μM, 0.5 Ci/mmol) or oligo(dT) (0.25 μM) and [α-32P]rATP (10 μM, 20 Ci/mmol) in the standard assay mixture. (B) Sizes of the products synthesized by SsoPriSL on oligo(dC) templates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with templates (0.25 μM) and [α-32P]rGTP (10 μM, 0.5 Ci/mmol) in the standard assay mixture. (C) Dependence of SsoPriSL activity on a dC stretch in template. SsoPriSL (0.5 μM) was incubated for 10 min at 55°C with templates (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. C34ddC was an oligo(dC) blocked at the 3′-end by a dideoxy-CMP. (D) Nuclease digestion analysis of products synthesized by SsoPriSL on oligo(dC). SsoPriSL (0.5 μM) was incubated for 10 min at 55°C with C35 (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. Samples were treated with indicated nucleases. (E) Nuclease digestion analysis of products synthesized by SsoPriSL on oligo(dT). SsoPriSL (1.5 μM) was incubated for 60 min at 55°C with T120 (0.25 μM) and [α-32P]rATP (10 μM, 10 Ci/mmol) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% (A, C, D and E) or 5% (B) polyacrylamide containing 7 M urea. (F) A diagram showing primer synthesis or terminal transfer at the 3′-end of the ssDNA template as the initial step in PADT.
Figure 2.
Influence of template sequences on PADT by SsoPriSL. (A) and (B) PADT on different templates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with an indicated template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rGTP (10 μM) in the standard assay mixture. (C) Synthesis by SsoPriSL on T32C3 with additional 1–3 dTs at the 3′-end. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with an indicated template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence of unlabeled rGTP (10 μM) in the standard assay mixture. (D) A diagram showing the proposed role of a dC stretch in a template in PADT. (E) Nuclease digestion analysis of products synthesized by SsoPriSL on T32C3 with [α-32P]rATP and unlabeled rGTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with T32C3 (0.25 μM), [α-32P]rATP (10 μM, 5 Ci/mmol) and unlabeled rGTP (10 μM) in the standard assay mixture. Samples were treated with indicated nucleases. In the RNase H + DNase I lane, the product was incubated sequentially with RNase H and DNase I. (F) Comparison of the PADT activities of SsoPriSL on templates containing dC and dG stretches of various sizes. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a template (0.25 μM) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the presence of unlabeled rGTP or rCTP (10 μM, for templates containing dCs or dGs, respectively) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea.
Figure 3.
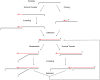
Sequencing analysis of PADT products synthesized by SsoPriSL. (A) Occurrences of indicated sequence stretches at the 5′-end of the PADT products. (B) Occurrences of rA stretches of different lengths and with different downstream nucleotides in the middle portion of the PADT products. (C) An interpretation of the sequencing results. Incorporated ribonucleotides are indicated by letters in lower case.
Figure 4.
SsoPriSL promoted annealing between strands with short regions of sequence complementarity and subsequent strand extension. (A) Annealing of oligonucleotides with short complementary ends and subsequent strand extension by SsoPriSL. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a specified pair of oligonucleotides (0.25 μM each) and [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) Nuclease digestion analysis of reaction products from T32C3/T32G3. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with T32C3 and T32G3 (0.25 μM each) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. Samples were treated with indicated nucleases. (C) A diagram depicting the optimal length of matching dC and dG sequences for SsoPriSL-promoted strand annealing. (D) Dependence of SsoPriSL-promoted strand annealing and subsequent extension on the length of the non-paring part of the strands. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a specified pair of oligonucleotides (0.25 μM each) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) in the standard assay mixture. (E) Effect of primer size on the use of a short template by SsoPriSL. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with a primer of different lengths (0.25 μM) and a short template (0.25 μM) in the presence of [α-32P]rUTP (10 μM, 5 Ci/mmol) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% (A, B) or 15% (D and E) polyacrylamide containing 7 M urea.
Figure 5.
Activities of SsoPriSL at 70°C. (A) Synthesis on oligo(dC). SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with a template (0.25 μM) and [α-32P]rGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) Terminal nucleotidyl transferase activity. SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with C59 (0.25 μM) and [α-32P]rATP (5 μM, 50 Ci/mmol) in the standard assay mixture. (C) PADT activity. SsoPriSL (0.5 μM) was incubated for 30 min at 70°C with a template (0.25 μM) in the presence of [α-32P]rATP (10 μM, 5 Ci/mmol) and unlabeled rGTP (10 μM) in the standard assay mixture. Reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea. Bands indicated by an arrow (B) are short products that SsoPriSL was able to synthesize in the absence of a template. These products have been shown to migrate with aberrantly slow mobility. For example, the lower band corresponds to a di-ribonucleotide (9).
Figure 6.
Activities of SsoPriSL with dNTPs as the substrates. (A) Synthesis on oligo(dC). SsoPriSL (0.5 μM) was incubated for 30 min at 55 or 70°C with C35 or C34ddC (0.25 μM) and [α-32P]dGTP (10 μM, 5 Ci/mmol) in the standard assay mixture. (B) PADT activity on oligo(dC) with [α-32P]dGTP and unlabeled rGTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with C35 (0.25 μM) and [α-32P]dGTP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rGTP (10 μM) in the standard assay mixture. (C) PADT activity on oligo(dG) with [α-32P]dCTP and unlabeled rCTP as the substrates. SsoPriSL (0.5 μM) was incubated for 30 min at 55°C with G35 (0.25 μM) and [α-32P]dCTP (10 μM, 5 Ci/mmol) in the presence or absence of unlabeled rCTP (10 μM) in the standard assay mixture. (D) and (E) PADT activity on various templates with dNTPs as the substrates. SsoPriSL (1.5 μM) was incubated for 30 min at 55°C with a specified template (0.25 μM) and [α-32P]dATP (1 μM, 50 Ci/mmol) in the presence or absence of unlabeled dGTP or dCTP (10 μM, for templates containing dCs or dGs, respectively) in the standard assay mixture. All reaction products were subjected to electrophoresis in 12% polyacrylamide containing 7 M urea. Bands indicated by an arrow (D and E) are short products that SsoPriSL was able to synthesize in the absence of a template.
Figure 7.
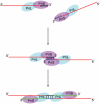
A model for PADT by SsoPriSL.
Figure 8.
A diagram showing the formation of a proposed ternary complex during SsoPriSL-promoted strand annealing.
Similar articles
- Interplay between primase and replication factor C in the hyperthermophilic archaeon Sulfolobus solfataricus.
Wu K, Lai X, Guo X, Hu J, Xiang X, Huang L. Wu K, et al. Mol Microbiol. 2007 Feb;63(3):826-37. doi: 10.1111/j.1365-2958.2006.05535.x. Epub 2006 Dec 20. Mol Microbiol. 2007. PMID: 17181784 - Characterization of a functional DnaG-type primase in archaea: implications for a dual-primase system.
Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA. Zuo Z, et al. J Mol Biol. 2010 Apr 2;397(3):664-76. doi: 10.1016/j.jmb.2010.01.057. Epub 2010 Feb 1. J Mol Biol. 2010. PMID: 20122937 - The chromosome replication machinery of the archaeon Sulfolobus solfataricus.
Duggin IG, Bell SD. Duggin IG, et al. J Biol Chem. 2006 Jun 2;281(22):15029-32. doi: 10.1074/jbc.R500029200. Epub 2006 Feb 8. J Biol Chem. 2006. PMID: 16467299 Review. - The Pol α-primase complex.
Pellegrini L. Pellegrini L. Subcell Biochem. 2012;62:157-69. doi: 10.1007/978-94-007-4572-8_9. Subcell Biochem. 2012. PMID: 22918585 Review.
Cited by
- PrimPol-A new polymerase on the block.
Rudd SG, Bianchi J, Doherty AJ. Rudd SG, et al. Mol Cell Oncol. 2014 Oct 29;1(2):e960754. doi: 10.4161/23723548.2014.960754. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308331 Free PMC article. Review. - An extended network of genomic maintenance in the archaeon Pyrococcus abyssi highlights unexpected associations between eucaryotic homologs.
Pluchon PF, Fouqueau T, Crezé C, Laurent S, Briffotaux J, Hogrel G, Palud A, Henneke G, Godfroy A, Hausner W, Thomm M, Nicolas J, Flament D. Pluchon PF, et al. PLoS One. 2013 Nov 7;8(11):e79707. doi: 10.1371/journal.pone.0079707. eCollection 2013. PLoS One. 2013. PMID: 24244547 Free PMC article. - Diversity of the DNA replication system in the Archaea domain.
Sarmiento F, Long F, Cann I, Whitman WB. Sarmiento F, et al. Archaea. 2014 Mar 26;2014:675946. doi: 10.1155/2014/675946. eCollection 2014. Archaea. 2014. PMID: 24790526 Free PMC article. Review. - Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes.
Guilliam TA, Keen BA, Brissett NC, Doherty AJ. Guilliam TA, et al. Nucleic Acids Res. 2015 Aug 18;43(14):6651-64. doi: 10.1093/nar/gkv625. Epub 2015 Jun 24. Nucleic Acids Res. 2015. PMID: 26109351 Free PMC article. Review.
References
- Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. - PubMed
- Rowen L, Kornberg A. A ribo-deoxyribonucleotide primer synthesized by primase. J. Biol. Chem. 1978;253:770–774. - PubMed
- Swart JR, Griep MA. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 1993;268:12970–12976. - PubMed
- Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–16106. - PubMed