Skin microbiome: looking back to move forward - PubMed (original) (raw)
Review
. 2012 Mar;132(3 Pt 2):933-9.
doi: 10.1038/jid.2011.417. Epub 2011 Dec 22.
Affiliations
- PMID: 22189793
- PMCID: PMC3279608
- DOI: 10.1038/jid.2011.417
Review
Skin microbiome: looking back to move forward
Heidi H Kong et al. J Invest Dermatol. 2012 Mar.
Abstract
Trillions of bacteria, fungi, viruses, archaea, and small arthropods colonize the skin surface, collectively comprising the skin microbiome. Generations of researchers have classified these microbes as transient versus resident, beneficial versus pathogenic, and collaborators versus adversaries. Culturing and direct sequencing of microbial inhabitants identified distinct populations present at skin surface sites. Herein, we explore the history of this field, describe findings from the current molecular sequencing era, and consider the future of investigating how microbes and antimicrobial therapy contribute to human health.
Figures
Figure 1. Schematic of skin histology viewed in cross-section with microorganisms and skin appendages
Microorganisms (virus, bacteria and fungi, mites) cover the surface of the skin and reside deep within the hair and glands.
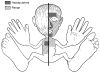
Figure 2. Homonculus showing major sites and range of Propionibacterium acnes colonization
Classic microbiologists swabbed and cultured diverse sites to characterize the predominant sites of colonization (“headquarters”) and general distribution (“range”). Adapted from Marples’ “The Ecology of the Human Skin”.
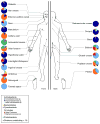
Figure 3. Topographical distribution of bacteria on skin sites
The skin microbiome is highly dependent on the microenvironment of the sampled site. Sebaceous sites are labeled in blue, moist sites are labeled in green and dry surfaces are labeled in red. Family-level classification of bacteria colonizing an individual subject is shown. Data is from Grice et al, 2009.
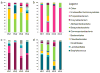
Figure 4. Interpersonal variation of the skin microbiome
Characterization of the skin microbiota, as determined by 16S rRNA sequencing, of four sites on four healthy volunteers (HV1, HV2, HV3, HV4) is depicted. Skin microbial variation is more similar dependent on the site than the individual. A. Antecubital crease; B. Back; C. Nare; D. Plantar heel. Data from Grice et al, 2009.
Similar articles
- The Skin Microbiome in Atopic Dermatitis and Its Relationship to Emollients.
Lynde CW, Andriessen A, Bertucci V, McCuaig C, Skotnicki S, Weinstein M, Wiseman M, Zip C. Lynde CW, et al. J Cutan Med Surg. 2016 Jan;20(1):21-8. doi: 10.1177/1203475415605498. Epub 2015 Oct 22. J Cutan Med Surg. 2016. PMID: 26492918 Review. - Skin Barrier Insights: From Bricks and Mortar to Molecules and Microbes.
Bosko CA. Bosko CA. J Drugs Dermatol. 2019 Jan 1;18(1s):s63-67. J Drugs Dermatol. 2019. PMID: 30681811 Review. - Skin disease modeling from a mathematical perspective.
Tanaka RJ, Ono M. Tanaka RJ, et al. J Invest Dermatol. 2013 Jun;133(6):1472-8. doi: 10.1038/jid.2013.69. Epub 2013 Mar 21. J Invest Dermatol. 2013. PMID: 23514928 - The skin microbiome: current perspectives and future challenges.
Chen YE, Tsao H. Chen YE, et al. J Am Acad Dermatol. 2013 Jul;69(1):143-55. doi: 10.1016/j.jaad.2013.01.016. Epub 2013 Mar 13. J Am Acad Dermatol. 2013. PMID: 23489584 Free PMC article. Review. - Atopic dermatitis microbiomes stratify into ecologic dermotypes enabling microbial virulence and disease severity.
Tay ASL, Li C, Nandi T, Chng KR, Andiappan AK, Mettu VS, de Cevins C, Ravikrishnan A, Dutertre CA, Wong XFCC, Ng AHQ, Matta SA, Ginhoux F, Rötzschke O, Chew FT, Tang MBY, Yew YW, Nagarajan N, Common JEA. Tay ASL, et al. J Allergy Clin Immunol. 2021 Apr;147(4):1329-1340. doi: 10.1016/j.jaci.2020.09.031. Epub 2020 Oct 8. J Allergy Clin Immunol. 2021. PMID: 33039480
Cited by
- Understanding and treating diabetic foot ulcers: Insights into the role of cutaneous microbiota and innovative therapies.
Norton P, Trus P, Wang F, Thornton MJ, Chang CY. Norton P, et al. Skin Health Dis. 2024 May 30;4(4):e399. doi: 10.1002/ski2.399. eCollection 2024 Aug. Skin Health Dis. 2024. PMID: 39104636 Free PMC article. Review. - The Role of Collagens in Atopic Dermatitis.
Szalus K, Trzeciak M. Szalus K, et al. Int J Mol Sci. 2024 Jul 12;25(14):7647. doi: 10.3390/ijms25147647. Int J Mol Sci. 2024. PMID: 39062889 Free PMC article. Review. - Impact of Bio-impedance Emitted from Wearable Smart Watches on Skin Microbiota: A Pilot Study.
Kumar DS, Sankar MM. Kumar DS, et al. J Pharm Bioallied Sci. 2024 Apr;16(Suppl 2):S1223-S1225. doi: 10.4103/jpbs.jpbs_546_23. Epub 2024 Apr 16. J Pharm Bioallied Sci. 2024. PMID: 38882903 Free PMC article. - Altered skin microbiome, inflammation, and JAK/STAT signaling in Southeast Asian ichthyosis patients.
Ho M, Nguyen HN, Van Hoang M, Bui TTT, Vu BQ, Dinh THT, Vo HTM, Blaydon DC, Eldirany SA, Bunick CG, Bui CB. Ho M, et al. Hum Genomics. 2024 Apr 16;18(1):38. doi: 10.1186/s40246-024-00603-x. Hum Genomics. 2024. PMID: 38627868 Free PMC article. - Characterization of the skin microbiome in normal and cutaneous squamous cell carcinoma affected cats and dogs.
Bromfield JI, Zaugg J, Straw RC, Cathie J, Krueger A, Sinha D, Chandra J, Hugenholtz P, Frazer IH. Bromfield JI, et al. mSphere. 2024 Apr 23;9(4):e0055523. doi: 10.1128/msphere.00555-23. Epub 2024 Mar 26. mSphere. 2024. PMID: 38530017 Free PMC article.
References
- Allen AM, Taplin D. Tropical immersion foot. Lancet. 1973;2:1185–9. - PubMed
- Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001;6:170–4. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources