Genome architectures revealed by tethered chromosome conformation capture and population-based modeling - PubMed (original) (raw)
Genome architectures revealed by tethered chromosome conformation capture and population-based modeling
Reza Kalhor et al. Nat Biotechnol. 2011.
Abstract
We describe tethered conformation capture (TCC), a method for genome-wide mapping of chromatin interactions. By performing ligations on solid substrates rather than in solution, TCC substantially enhances the signal-to-noise ratio, thereby facilitating a detailed analysis of interactions within and between chromosomes. We identified a group of regions in each chromosome in human cells that account for the majority of interchromosomal interactions. These regions are marked by high transcriptional activity, suggesting that their interactions are mediated by transcriptional machinery. Each of these regions interacts with numerous other such regions throughout the genome in an indiscriminate fashion, partly driven by the accessibility of the partners. As a different combination of interactions is likely present in different cells, we developed a computational method to translate the TCC data into physical chromatin contacts in a population of three-dimensional genome structures. Statistical analysis of the resulting population demonstrates that the indiscriminate properties of interchromosomal interactions are consistent with the well-known architectural features of the human genome.
Figures
Figure 1. Overview of Tethered Conformation Capture (TCC)
Cells are treated with formaldehyde, which covalently crosslinks proteins (purple ellipses) to each other and to DNA (orange and blue strings). (1) The chromatin is solubilized and its proteins are biotinylated (purple ball and stick). DNA is digested with a restriction enzyme that generates 5’ overhangs. (2) Crosslinked complexes are immobilized at a very low density on the surface of streptavidin coated magnetic beads (grey arc) through the biotinylated proteins; non-crosslinked DNA fragments are removed. (3) The 5’ overhangs are filled in with an α-thio-triphosphate containing nucleotide analog (the yellow nucleotide in the inset), which is resistant to exonuclease digestion, and a biotinylated nucleotide analog (the red nucleotide with the purple ball and stick in the inset) to generate blunt ends. (4) Blunt DNA ends are ligated. (5) Crosslinking is reversed and DNA is purified. The biotinylated nucleotide is removed from non-ligated DNA ends using E. coli exonuclease III while the phosphorothioate bond protects DNA fragments from complete degradation. (6) The DNA is sheared and fragments that include a ligation junction are isolated on streptavidin-coated magnetic beads, but this time through the biotinylated nucleotides. (7) Sequencing adaptors are added to all DNA molecules to generate a library. (8) Ligation events are identified using paired-end sequencing. The steps that are unique to the TCC strategy are biotinylation of the chromatin proteins, immobilization of crosslinked complexes on the beads, performing ligation and other reactions on the beads, and the use of exonuclease-resistance nucleotide analogs for the purification of ligated DNA fragments from the non-ligated.
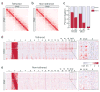
Figure 2. Tethering improves the signal-to-noise ratio of conformation capture
(a,b) TCC can reproduce the results obtained by Hi-C. A genome-wide contact frequency map is compiled from the ligation frequency data generated by tethered (TCC) (a) and non-tethered (Hi-C) (b) conformation capture. The portion of each map that corresponds to the intra-chromosomal contacts of chromosome 2 is shown. The intensity of the red color in each position of the map represents the observed frequency of contact between corresponding segments of the chromosome which are shown on the top and to the left of the map. In these maps, chromosomes 2 is divided into segments that span 277 HindIII sites each, resulting in 258 segments of ∼1 Mb (Supplementary Methods). A pair of tick marks on the ideogram encompasses 4986 HindIII sites. In this and other figures, the white lines in the heatmaps mark the unalignable region of the centromeres. See Supplementary Figure 1a for the tethered contact frequency maps of all the other chromosomes. (c) The observed fractions of intra (dark red) and inter-chromosomal (light blue) ligations in tethered (T) and non-tethered (NT) libraries produced using HindIII or MboI. The random ligation (RL) bar represents the expected fractions if all ligations occurred between non-crosslinked DNA fragments. For the non-tethered MboI library only, these fractions were determined by sequencing 160 individual DNA molecules from three replicates of the experiment. See Supplementary Table 1 for the sequencing output information of the other three libraries. (d,e) The genome-wide enrichment map for chromosome 2, compiled from the tethered (d) and non-tethered (e) HindIII libraries. Enrichment is calculated as the ratio of the observed frequency in each position to its expected value; expected values were obtained assuming completely random ligations (Methods). Red and light blue respectively indicate enrichment and depletion of a contact in accordance with the color key between the panels. Chromosome 2 (left) extends along the Y-axis while all 23 chromosomes (top) extend along the X-axis. The zoomed panel to the right of each map magnifies the section that corresponds to contacts between the small arm of chromosome 2 and chromosomes 20, 21, 22, and X. For these maps, each chromosome is divided into segments that span 558 HindIII sites, leading to respectively 116 and 1384 segments of ∼1.5 Mb for chromosome 2 and all other chromosomes. A pair of tick marks on chromosome 2 spans 5022 HindIII sites.
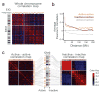
Figure 3. Intra-chromosomal interactions
(a) Correlation map and class assignment for chromosome 2. The color of each position in the map represents the Pearson’s correlation between the intra-chromosomal contact profiles of the corresponding two segments of the chromosome to the left and on top (the ideogram of the chromosome has only been shown to the left, but the X-axis of the map also represents the chromosome). The color key is shown on the bottom-right corner of the figure. To assign each segment to the active (orange blocks on top of the map) or the inactive (purple blocks on top of the map) class, principal component analysis (PCA) is used to calculate the EIG variable (plotted on top of the assignment blocks) for each segment. Segments with a positive EIG are assigned to the active class, while those with a negative EIG are assigned to the inactive class. Segments with EIG values close to zero have not been assigned to either class (Methods). The size of each chromosome band is based on the number of HindIII sites it contains. For this map, chromosome 2 is divided into 517 segments of ∼0.5 Mb, each spanning 138 HindIII sites. See Supplementary Figure 2a for correlation maps and class assignments of all the other autosomal chromosomes. Data from the tethered HindIII library are used in this panel and other panels of the figure. (b) The genome-wide average Pearson’s correlation between intra-chromosomal contact profiles of two active segments (orange), two inactive segments (dark purple), and an active and an inactive segment (gray) plotted against their genomic distance. Each chromosome is divided into segments of 138 HindIII sites, resulting in 6,000 segments of ∼0.5 Mb. (c) Active-active (left) and inactive-inactive (right) correlation maps for chromosome 2. The color intensity of each point in the map represents the Pearson’s correlation between the “active-only” (left) or “inactive-only” (right) contact profiles of the corresponding segments, whose location in the chromosome has been marked by an arrow on the ideogram of chromosome 2 in the middle. The ideogram shows the positions of the active (orange bars on the left) and inactive (purple bars on the right) segments. The different shades of orange and purple are used only to differentiate the adjacent segments. Each correlation map is calculated following the procedure in (a), except only contacts between active segments (left) or inactive segments (right) are considered. Color-coding is identical to (a) and the key is shown on the bottom-right corner of the figure. The order of segments from left to right is the same as the order from top to bottom. Segment sizes are identical to (a). For similar active-active and inactive-inactive maps of the other large chromosomes see Supplementary Figure 3c.
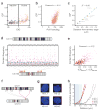
Figure 4. Inter-chromosomal interactions
(a) For all segments of chromosome 2, inter-chromosomal contact probability index (ICP) is plotted against EIG. Segments with a positive EIG (orange) belong to the active class, while those with a negative EIG (brown) belong to the inactive class. The blue dashed line separates high-ICP segments: values above the line are significantly larger than the average ICP for inactive segments. Red dots mark those inactive segments with a large ICP that also flank the centromere. For this map, chromosome 2 is divided into 517 segments of ∼0.5 Mb, each spanning 138 HindIII sites. See Supplementary Figure 4a for similar plots of all autosomal chromosomes and Supplementary Figure 4b for the alignment of ICP and EIG values along chromosome 2. See also Supplementary Table 2 for ICP and EIG values of all segments of the genome. In this and other panels of this figure, data from the tethered HindIII library are used. (b) For all active segments in the genome, ICP is plotted against the binding of RNA polymerase II (pol II). Pol II binding values are reproduced from a ChIP-seq study on the GM12878 cells and are in arbitrary units based on alignment frequency (Methods). The p-value of the correlation is smaller than 10-16. Each point represents a segment of the genome that spans 138 HindIII sites. The X-axis is plotted in a logarithmic scale. (c) For seven loci on the small arm of chromosome 11, the ICP value is plotted against their average distance from the edge of chromosome 11 territory as measured by FISH. Positive distance values denote localization within the bulk territory, while negative values denote localization away from the bulk territory. Orange and brown dots represent assignment to the active and inactive classes respectively. Error bars represent ±95% confidence interval. See Supplementary Figure 4c for more information and a side-by-side comparison of the FISH and TCC data for these loci. (d) Plotted are the frequencies of all contacts between high-ICP active segments on chromosome 19 and all the segments on chromosome 11. Contacts involving high-ICP active segments on chromosome 11 are shown as purple squares and contacts involving all other segments of this chromosome are shown as grey triangles. Contacts plotted between vertical dotted lines involve the same high-ICP active segment on chromosome 19 and all the segments of chromosome 11. Frequencies above the dashed blue line are significantly higher than the average frequency of contacts between high-ICP active segments on chromosome 19 and inactive segments on chromosome 11 (p-value < 0.04, non-parametric). These frequencies can be considered significantly larger than the noise level, defined as the false-positive contact frequencies due to random inter-molecular ligations. For this plot, each chromosome was divided into ∼1 Mb segments that span 277 HindIII sites resulting in a total of 143 segments for chromosomes 11 and 43 segments for chromosome 19. Among those, 14 segments on chromosome 19 and 28 segments on chromosome 11 were classified as high-ICP active. The locations of the high-ICP active segments in chromosome 19 are marked by an orange bar on the ideogram of the chromosome on the bottom of the panel. The different shades of oranges are used only to differentiate the adjacent segments. See Supplementary Figure 6a for contact profiles of high-ICP active segments in chromosome 19 with all high-ICP active segments in the genome. (e) For all possible pairs of high-ICP active segments from chromosomes 11 and 19, their contact frequency has been plotted against the product of their _ICP_s. Same interactions are marked with purple color in (d). The p-value of the correlation is nominal.Other parameters are the same as in (d). See also Supplementary Figure 6b for a similar plot of chromosome 11 with all the other chromosomes and Supplementary Figure 6c for a histogram of the correlations of all 231 possible such plots for autosomal chromosomes. (f) The layout of 3D-FISH experiments where the localization of a high-ICP active locus on chromosome 19 (H0) relative to four loci on chromosome 11 (H1, H2, L1, and L2) was analyzed in about 1,000 cells per pair of loci. H1 and H2 are high-ICP active, while the L1 and L2 are inactive. The blocks on the chromosomes' ideograms mark the position of each locus (orange for high-ICP active and brown for inactive), and the arrows mark the pair combinations that are analyzed (purple for active-active and grey for active-inactive). See also Supplementary Table 3 for the names and genomic locations of the BAC clones that were used. (g) An example nucleus from each pair of loci analyzed in 3D-FISH. Nuclei are counterstained with DAPI (blue). In all four nuclei, the hybridization signal of H0 is shown in red and that of the other locus is shown in green. (h) Cumulative percentage of nuclei that show a pair of hybridization signals closer than a given distance is plotted. Only the closest pair of signals for each nucleus is considered. 1,011, 987, 976, and 998 total nuclei were analyzed in duplicates for H0-L1, H0-L2, H0-H1, and H0-H2 respectively. Distances smaller than 0.6 μm (dashed blue line - arbitrarily selected for visualization purposes) represent colocalizations in a close vicinity where a direct interaction between loci is possible. Because colocalization is required but not sufficient for a direct contact, these values likely provide a ceiling for the fraction of cells that harbor a direct contact between these loci.
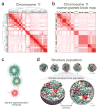
Figure 5. Coarse-graining of the contact frequency maps and structural representation of the genome
(a) The contact frequency map of chromosome 11 from the tethered HindIII library. The chromosome has been divided into 237 segments each of which covers 166 HindIII sites. Hierarchical constrained clustering was applied using the Pearson’s correlation between the segments’ contact profiles as the similarity measure (Methods). The dendrogram of constrained clustering is shown to the left and on top of the map. The intensity of the red color in each position of the map represents the observed frequency of contact between corresponding segments of the chromosome shown on the top and to the left of the map. (b) Coarse-grained block matrix of chromosome 11. To identify the blocks, a clustering cutoff was determined following a previously described procedure. In the block map, the value of an element is the average contact frequency of all the corresponding elements in the contact frequency map. The dimension of the initial contact frequency map is reduced to 15 blocks for chromosome 11 and 428 for the entire genome in the block map. Spearman’s rank correlation coefficient between this block matrix and the contact frequency map in (a) is 0.78. Assignment of segments to the active (orange blocks) and inactive (dark brown blocks) classes are shown to the left and on top of the matrix. The intensity of the red color in each element represents the average of the observed contact frequencies between the corresponding blocks of the chromosome. See Supplementary Figure 7b-d for the coarse-grained genome-wide block matrix. (c) Sphere representation for chromatin regions in a block. The sphere for each block is defined by two different radii. First, its hard radius (solid sphere) which is estimated from the block sequence length and nuclear occupancy of the genome; the sphere cannot be penetrated within this radius (Methods). Second, its soft radius (dotted line), which is twice that of the hard sphere radius. A contact between two spheres is defined as an overlap between the spheres’ respective soft radii. Also shown is a schematic hypothetical view of the chromatin fiber. For all the block sequence lengths and resulting sphere radii see Supplementary Table 4. (d) Genome structure population of 10,000. A schematic of the calculated structure population is shown on top. A randomly selected sample from the population is magnified on the buttom. All forty-six chromosome territories are shown. Homologous pairs share the same color. The nuclear envelope is displayed in grey. For visualization purposes, the spheres are blurred in the magnified structure because the use of 2x428 spheres to represent the genome makes the territories appear more discrete than they actually are.
Figure 6. Population-based analysis of territory localizations in the nucleus
(a) The distribution of the radial positions for chromosomes 18 (red dashed line) and 19 (blue solid line), calculated from the genome structure population. Radial positions are calculated for the center of mass of each chromosome and are given as a fraction of the nuclear radius. See Supplementary Figure 9b for the radial distribution of all chromosome territories. (b) The average radial position of all chromosomes plotted against their size. Error bars mark the standard deviation. For the radial positions from a control genome structure population generated without TCC data see Supplementary Figure 9a. (c) Clustering of chromosomes with respect to the average distance between the center of mass of each chromosome pair in the genome structure population (shorter to longer average distance is colored by gradual purple to white). The clustering dendogram, which identifies two clusters is shown on top. (d) (Left panels) The density contour plot of the localization probability for all chromosomes in cluster 1 (top panel) and cluster 2 (bottom panel) calculated from all the structures in the genome structure population. The rainbow color-coding ranges from blue (minimum value) to red (maximum value). (Right panels) Shown is a representative genome structure from the genome structure population. Chromosome territories are shown for all chromosomes in cluster 1 (top) and all chromosomes in clusters 2 (bottom). The localization probabilities are calculated following a previously-described procedure.
Comment in
- Parallel genome universes.
Misteli T. Misteli T. Nat Biotechnol. 2012 Jan 9;30(1):55-6. doi: 10.1038/nbt.2085. Nat Biotechnol. 2012. PMID: 22231096 Free PMC article. No abstract available.
Similar articles
- Population-based 3D genome structure analysis reveals driving forces in spatial genome organization.
Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, Zhou Y, Li H, Zhou XJ, Le Gros MA, Larabell CA, Chen L, Alber F. Tjong H, et al. Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):E1663-72. doi: 10.1073/pnas.1512577113. Epub 2016 Mar 7. Proc Natl Acad Sci U S A. 2016. PMID: 26951677 Free PMC article. - Parallel genome universes.
Misteli T. Misteli T. Nat Biotechnol. 2012 Jan 9;30(1):55-6. doi: 10.1038/nbt.2085. Nat Biotechnol. 2012. PMID: 22231096 Free PMC article. No abstract available. - A Guided Protocol for Array Based T2C: A High-Quality Selective High-Resolution High-Throughput Chromosome Interaction Capture.
Knoch TA. Knoch TA. Curr Protoc Hum Genet. 2018 Oct;99(1):e55. doi: 10.1002/cphg.55. Epub 2018 Sep 10. Curr Protoc Hum Genet. 2018. PMID: 30199150 - Chromatin and epigenetic features of long-range gene regulation.
Harmston N, Lenhard B. Harmston N, et al. Nucleic Acids Res. 2013 Aug;41(15):7185-99. doi: 10.1093/nar/gkt499. Epub 2013 Jun 13. Nucleic Acids Res. 2013. PMID: 23766291 Free PMC article. Review.
Cited by
- A streamlined tethered chromosome conformation capture protocol.
Gabdank I, Ramakrishnan S, Villeneuve AM, Fire AZ. Gabdank I, et al. BMC Genomics. 2016 Apr 1;17:274. doi: 10.1186/s12864-016-2596-3. BMC Genomics. 2016. PMID: 27036078 Free PMC article. - Identification of genes associated with the astrocyte-specific gene Gfap during astrocyte differentiation.
Ito K, Sanosaka T, Igarashi K, Ideta-Otsuka M, Aizawa A, Uosaki Y, Noguchi A, Arakawa H, Nakashima K, Takizawa T. Ito K, et al. Sci Rep. 2016 Apr 4;6:23903. doi: 10.1038/srep23903. Sci Rep. 2016. PMID: 27041678 Free PMC article. - 4D nucleomes in single cells: what can computational modeling reveal about spatial chromatin conformation?
Sekelja M, Paulsen J, Collas P. Sekelja M, et al. Genome Biol. 2016 Apr 7;17:54. doi: 10.1186/s13059-016-0923-2. Genome Biol. 2016. PMID: 27052789 Free PMC article. Review. - Closing the loop: 3C versus DNA FISH.
Giorgetti L, Heard E. Giorgetti L, et al. Genome Biol. 2016 Oct 19;17(1):215. doi: 10.1186/s13059-016-1081-2. Genome Biol. 2016. PMID: 27760553 Free PMC article. Review. - Physical mechanisms behind the large scale features of chromatin organization.
Pombo A, Nicodemi M. Pombo A, et al. Transcription. 2014;5(2):e28447. doi: 10.4161/trns.28447. Transcription. 2014. PMID: 25764220 Free PMC article. Review.
References
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
- Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127–134. - PubMed
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
- Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. - PubMed
- Cremer M, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR022220/RR/NCRR NIH HHS/United States
- R01 GM096089/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 GM077320/GM/NIGMS NIH HHS/United States
- GM064642/GM/NIGMS NIH HHS/United States
- GM096089/GM/NIGMS NIH HHS/United States
- R01 GM064642/GM/NIGMS NIH HHS/United States
- GM077320/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials