The interface of transcription and DNA replication in the mitochondria - PubMed (original) (raw)
Review
The interface of transcription and DNA replication in the mitochondria
Rajesh Kasiviswanathan et al. Biochim Biophys Acta. 2012 Sep-Oct.
Abstract
DNA replication of the mitochondrial genome is unique in that replication is not primed by RNA derived from dedicated primases, but instead by extension of processed RNA transcripts laid down by the mitochondrial RNA polymerase. Thus, the RNA polymerase serves not only to generate the transcripts but also the primers needed for mitochondrial DNA replication. The interface between this transcription and DNA replication is not well understood but must be highly regulated and coordinated to carry out both mitochondrial DNA replication and transcription. This review focuses on the extension of RNA primers for DNA replication by the replication machinery and summarizes the current models of DNA replication in mitochondria as well as the proteins involved in mitochondrial DNA replication, namely, the DNA polymerase γ and its accessory subunit, the mitochondrial DNA helicase, the single-stranded DNA binding protein, topoisomerase I and IIIα and RNaseH1. This article is part of a Special Issue entitled: Mitochondrial Gene Expression.
Published by Elsevier B.V.
Figures
Figure 1. Schematic diagram of a mitochondrial DNA replication fork showing the critical proteins required for DNA replication
The nascent DNA synthesized by pol γ (green) is shown as a solid red line, while the RNA primer (jagged red line) created by the mitochondrial RNA polymerase (orange) is being degraded by RNase H1 (yellow). The mitochondrial DNA helicase (purple) unwinds the downstream DNA forming a single-stranded loop which is coated with mtSSB (light blue). Topoisomerases (brown) work to relieve torsional tension in the DNA created by unwinding.
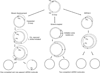
Figure 2. Models of mtDNA replication
Left panel. The asymmetric or strand displacement model. Replication of the H-strand is initiated at OriH with accompanying displacement of the H-strand thus forming a D-loop. This synthesis proceeds until OriL is exposed where synthesis of the L-stand is initiated in the opposite direction. Middle panel. The strand-coupled model. Bidirectional replication is initiated from a zone near OriH followed by progression of the two forks around the mtDNA circle. Right panel. The RITOLS model. Replication of the leading strand initiates similar to the strand-displacment model but the lagging strand is initially transcribed as RNA (dashed line) before being converted to DNA.
Similar articles
- In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication.
Miralles Fusté J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson Ö, Sabouri N, Gustafsson CM, Falkenberg M. Miralles Fusté J, et al. PLoS Genet. 2014 Dec 4;10(12):e1004832. doi: 10.1371/journal.pgen.1004832. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25474639 Free PMC article. - DNA replication and transcription in mammalian mitochondria.
Falkenberg M, Larsson NG, Gustafsson CM. Falkenberg M, et al. Annu Rev Biochem. 2007;76:679-99. doi: 10.1146/annurev.biochem.76.060305.152028. Annu Rev Biochem. 2007. PMID: 17408359 Review. - The human mitochondrial replication fork in health and disease.
Wanrooij S, Falkenberg M. Wanrooij S, et al. Biochim Biophys Acta. 2010 Aug;1797(8):1378-88. doi: 10.1016/j.bbabio.2010.04.015. Epub 2010 Apr 22. Biochim Biophys Acta. 2010. PMID: 20417176 Review. - Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro.
Wanrooij S, Fusté JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Wanrooij S, et al. Proc Natl Acad Sci U S A. 2008 Aug 12;105(32):11122-7. doi: 10.1073/pnas.0805399105. Epub 2008 Aug 6. Proc Natl Acad Sci U S A. 2008. PMID: 18685103 Free PMC article. - Purification and Comparative Assay of Human Mitochondrial Single-Stranded DNA-Binding Protein.
Ciesielski GL, Rosado-Ruiz FA, Kaguni LS. Ciesielski GL, et al. Methods Mol Biol. 2016;1351:211-22. doi: 10.1007/978-1-4939-3040-1_16. Methods Mol Biol. 2016. PMID: 26530685 Free PMC article.
Cited by
- Mitochondrial DNA: Consensuses and Controversies.
Shokolenko I, Alexeyev M. Shokolenko I, et al. DNA (Basel). 2022 Jun;2(2):131-148. doi: 10.3390/dna2020010. Epub 2022 Jun 10. DNA (Basel). 2022. PMID: 36381197 Free PMC article. - Protecting the mitochondrial powerhouse.
Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM 3rd, Bohr VA. Scheibye-Knudsen M, et al. Trends Cell Biol. 2015 Mar;25(3):158-70. doi: 10.1016/j.tcb.2014.11.002. Epub 2014 Dec 11. Trends Cell Biol. 2015. PMID: 25499735 Free PMC article. Review. - Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans.
Leung MC, Rooney JP, Ryde IT, Bernal AJ, Bess AS, Crocker TL, Ji AQ, Meyer JN. Leung MC, et al. BMC Pharmacol Toxicol. 2013 Feb 4;14:9. doi: 10.1186/2050-6511-14-9. BMC Pharmacol Toxicol. 2013. PMID: 23374645 Free PMC article. - Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae.
Moriyama T, Tajima N, Sekine K, Sato N. Moriyama T, et al. Genome Biol Evol. 2014 Jan;6(1):228-37. doi: 10.1093/gbe/evu009. Genome Biol Evol. 2014. PMID: 24407855 Free PMC article. - Regulation of the human Suv3 helicase on DNA by inorganic cofactors.
Venø ST, Witt MB, Kulikowicz T, Bohr VA, Stevnsner T. Venø ST, et al. Biochimie. 2015 Jan;108:160-8. doi: 10.1016/j.biochi.2014.11.003. Epub 2014 Nov 21. Biochimie. 2015. PMID: 25446650 Free PMC article.
References
- Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13534–13539. - PMC - PubMed
- Bogenhagen DF, Clayton DA. Concluding remarks: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:404–405. - PubMed
- Bogenhagen DF, Clayton DA. The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:357–360. - PubMed
- Holt IJ, Jacobs HT. Response: The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:355–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ES065078/ES/NIEHS NIH HHS/United States
- Z01 ES065080/Intramural NIH HHS/United States
- ZIA ES065078-18/Intramural NIH HHS/United States
- Z01 ES065078/Intramural NIH HHS/United States
- ZIA ES065080-17/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources