Descendants of primed Arabidopsis plants exhibit resistance to biotic stress - PubMed (original) (raw)
Descendants of primed Arabidopsis plants exhibit resistance to biotic stress
Ana Slaughter et al. Plant Physiol. 2012 Feb.
Abstract
An attack of plants by pathogens or treatment with certain resistance-inducing compounds can lead to the establishment of a unique primed state of defense. Primed plants show enhanced defense reactions upon further challenge with biotic or abiotic stress. Here, we report that the primed state in Arabidopsis (Arabidopsis thaliana) is still functional in the next generation without additional treatment. We compared the reactions of Arabidopsis plants that had been either primed with β-amino-butyric acid (BABA) or with an avirulent isolate of the bacteria Pseudomonas syringae pv tomato (PstavrRpt2). The descendants of primed plants showed a faster and higher accumulation of transcripts of defense-related genes in the salicylic acid signaling pathway and enhanced disease resistance upon challenge inoculation with a virulent isolate of P. syringae. In addition, the progeny of primed plants was also more resistant against the oomycete pathogen Hyaloperonospora arabidopsidis. When transgenerationally primed plants were subjected to an additional priming treatment, their descendants displayed an even stronger primed phenotype, suggesting that plants can inherit a sensitization for the priming phenomenon. Interestingly, this primed to be primed phenotype was much reduced in the Arabidopsis β-amino-butyric acid priming mutant ibs1 (induced BABA sterility1). Our results demonstrate that the primed state of plants is transferred to their progeny and confers improved protection from pathogen attack as compared to the descendants of unprimed plants.
Figures
Figure 1.
Experimental design. Parent plants: Ws-0 (Ws wild-type plants), ibs1 (a priming mutant in Ws-0 background), and Col-0 (Col wild-type plants). S1 progeny: descendants from the selfed parent plants; S2 progeny: descendants from the selfed S1 plants. The plants were treated with: water = H; BABA = B; mock treated with buffer = M; or inoculated with avirulent Pst pv tomato containing avrRpt2 = P.
Figure 2.
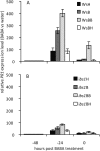
Basal PR1 expression levels in the progeny of Ws-0 plants and ibs1 plants (see Fig. 1). Four-week-old plants were soil drenched with water or a solution of BABA to a final concentration of 25 ppm in the soil. Transcript levels were analyzed with qRT-PCR. Expression was normalized to the sample treated with water at 0 h. PR1 expression in the different Ws-0 lines (A) and in the different ibs1 lines (B) is shown. The values represent means ±
sd
of three replicates. Similar results were obtained in three independent experiments.
Figure 3.
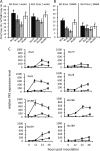
Descendants of BABA-treated Ws-0 plants are more resistant to virulent Pst. Three-week-old plants were treated with BABA (25 ppm final concentration in the soil) or water 2 d prior to inoculation with Pst DC3000 (OD600 = 0.08). A, Growth of Pst DC3000 in the water-treated Ws-0 lines (WsH, WsB, WsBB, and WsBH) and ibs1 lines (_ibs1_H, _ibs1_B, _ibs1_BB, and _ibs1_BH) at 72 hpi. B, Growth of Pst DC3000 in BABA-treated Ws-0 lines and in ibs1 lines at 72 h postinoculation. Bacterial growth was quantified by qRT-PCR as transcript levels of Psorf normalized to the transcript level of the Arabidopsis gene AtTUB4. Capital letters indicate statistically significant bacterial growth within the water-treated Ws-0 and ibs1 lines (ANOVA, Student-Newman-Keuls, n = 3, P < 0.05). Small letters indicate statistically significant bacterial growth within the BABA-treated Ws-0 or ibs1 lines. C, qRT-PCR analysis of PR1 gene expression in BABA-treated (circles) and water-treated (squares) Ws-0 and ibs1 lines. Expression was normalized to the corresponding sample treated with water at 0 h. The values represent means ±
sd
of three replicates. Similar results were obtained in three independent experiments.
Figure 4.
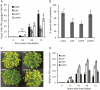
Inheritance of priming induced by avirulent Pst (PstavrRpt2). Single leaves of 4-week-old plants were infiltrated with Pst avrRpt2 or mock infiltrated with buffer. A, Col-0 lines (see Fig. 1) were challenge inoculated with Pst DC3000 and bacterial growth was quantified by qRT-PCR as described for Figure 3. Small and capital letters above error bars represent statistically significant difference in bacterial growth within Col-0 lines at 48 and 72 h postinoculation (ANOVA, Student-Newman-Keuls, n = 3, P < 0.001). Values represent means ±
sd
of three biological replicates. B, Quantification of disease resistance of Col lines 5 d post inoculation. Letters represent statistically significant difference in the percentage of leaves with symptoms (ANOVA, Student-Newman-Keuls, n = 30, P < 0.05). C, Disease phenotype of Col lines. D, qRT-PCR analysis of PR1 gene expression in Col lines. Expression was normalized to the sample treated with water at 0 h. The values represent means ±
sd
of three replicates. Similar results were obtained in three independent experiments.
Figure 5.
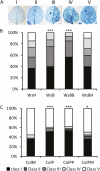
Inheritance of priming against H. arabidopsidis. A, Disease ratings were classified based on the percentage of leaf colonization by mycelium. Class I: 0%; class II: 1% to 25%; class III: 26% to 50%; class IV: 51% to 75%; and class V: 76% to 100%. Disease ratings are shown as the percentage of leaves in each resistance class. B, Disease resistance in Ws-0 lines inoculated with the Emwa strain of H. arabidopsidis. Asterisks indicate statistically significant differences in the percentage of leaves in the different classes compared to WsH (Fisher test, *** = P < 0.001). C, Disease resistance in Col lines inoculated with the Noco strain of H. arabidopsidis. Asterisks indicate statistically significant differences in the percentage of leaves in the different classes compared to ColM (Fisher test, * = P < 0.05; *** = P < 0.001).
Comment in
- Prime time for transgenerational defense.
Pieterse CM. Pieterse CM. Plant Physiol. 2012 Feb;158(2):545. doi: 10.1104/pp.112.900430. Plant Physiol. 2012. PMID: 22308198 Free PMC article. No abstract available.
Similar articles
- Next-generation systemic acquired resistance.
Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. Luna E, et al. Plant Physiol. 2012 Feb;158(2):844-53. doi: 10.1104/pp.111.187468. Epub 2011 Dec 5. Plant Physiol. 2012. PMID: 22147520 Free PMC article. - Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis.
Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Métraux JP, Mauch-Mani B. Ton J, et al. Plant Cell. 2005 Mar;17(3):987-99. doi: 10.1105/tpc.104.029728. Epub 2005 Feb 18. Plant Cell. 2005. PMID: 15722464 Free PMC article. - Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana.
Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CMJ. Pozo MJ, et al. New Phytol. 2008;180(2):511-523. doi: 10.1111/j.1469-8137.2008.02578.x. Epub 2008 Jul 24. New Phytol. 2008. PMID: 18657213 - Chemical priming of plant defense responses to pathogen attacks.
Hönig M, Roeber VM, Schmülling T, Cortleven A. Hönig M, et al. Front Plant Sci. 2023 May 8;14:1146577. doi: 10.3389/fpls.2023.1146577. eCollection 2023. Front Plant Sci. 2023. PMID: 37223806 Free PMC article. Review. - Ecological role of transgenerational resistance against biotic threats.
Rasmann S, De Vos M, Jander G. Rasmann S, et al. Plant Signal Behav. 2012 Apr;7(4):447-9. doi: 10.4161/psb.19525. Epub 2012 Apr 1. Plant Signal Behav. 2012. PMID: 22499174 Free PMC article. Review.
Cited by
- Epigenetic responses to stress: triple defense?
Gutzat R, Mittelsten Scheid O. Gutzat R, et al. Curr Opin Plant Biol. 2012 Nov;15(5):568-73. doi: 10.1016/j.pbi.2012.08.007. Epub 2012 Sep 7. Curr Opin Plant Biol. 2012. PMID: 22960026 Free PMC article. Review. - Next-generation systemic acquired resistance.
Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. Luna E, et al. Plant Physiol. 2012 Feb;158(2):844-53. doi: 10.1104/pp.111.187468. Epub 2011 Dec 5. Plant Physiol. 2012. PMID: 22147520 Free PMC article. - Effect of elicitors on holm oak somatic embryo development and efficacy inducing tolerance to Phytophthora cinnamomi.
Morcillo M, Sales E, Ponce L, Guillén A, Segura J, Arrillaga I. Morcillo M, et al. Sci Rep. 2020 Sep 16;10(1):15166. doi: 10.1038/s41598-020-71985-w. Sci Rep. 2020. PMID: 32938968 Free PMC article. - Comparative transcriptome expression analysis in susceptible and resistant potato (Solanum tuberosum) cultivars to common scab (Streptomyces scabies) revealed immune priming responses in the incompatible interaction.
Fofana B, Somalraju A, Fillmore S, Zaidi M, Main D, Ghose K. Fofana B, et al. PLoS One. 2020 Jul 16;15(7):e0235018. doi: 10.1371/journal.pone.0235018. eCollection 2020. PLoS One. 2020. PMID: 32673321 Free PMC article. - Epialleles in plant evolution.
Weigel D, Colot V. Weigel D, et al. Genome Biol. 2012 Oct 11;13(10):249. doi: 10.1186/gb-2012-13-10-249. Genome Biol. 2012. PMID: 23058244 Free PMC article. Review.
References
- Agrawal AA. (2002) Maternal effects associated with herbivory: mechanisms and consequences of transgenerational induced plant resistance. Ecology 83: 3408–3415
- Alvarez-Venegas R, Sadder M, Hlavacka A, Baluška F, Xia Y, Lu G, Firsov A, Sarath G, Moriyama H, Dubrovsky JG, et al. (2006) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci USA 103: 6049–6054 - PMC - PubMed
- Boachon B, Robert J, Daniel X, Mauch-Mani B. (2009) Absolute quantification of specific plant defence pathways and disease development of several microbial pathogens in Arabidopsis using real-time PCR. IOBC Meeting Proceedings of “Biological Control of Fungal and Bacterial Plant Pathogens (Phytopathogens)” Molecular Tools for Understanding and Improving Biocontrol, 43, pp 311–316
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases