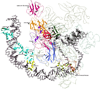Insights from the architecture of the bacterial transcription apparatus - PubMed (original) (raw)
Insights from the architecture of the bacterial transcription apparatus
Lakshminarayan M Iyer et al. J Struct Biol. 2012 Sep.
Abstract
We provide a portrait of the bacterial transcription apparatus in light of the data emerging from structural studies, sequence analysis and comparative genomics to bring out important but underappreciated features. We first describe the key structural highlights and evolutionary implications emerging from comparison of the cellular RNA polymerase subunits with the RNA-dependent RNA polymerase involved in RNAi in eukaryotes and their homologs from newly identified bacterial selfish elements. We describe some previously unnoticed domains and the possible evolutionary stages leading to the RNA polymerases of extant life forms. We then present the case for the ancient orthology of the basal transcription factors, the sigma factor and TFIIB, in the bacterial and the archaeo-eukaryotic lineages. We also present a synopsis of the structural and architectural taxonomy of specific transcription factors and their genome-scale demography. In this context, we present certain notable deviations from the otherwise invariant proteome-wide trends in transcription factor distribution and use it to predict the presence of an unusual lineage-specifically expanded signaling system in certain firmicutes like Paenibacillus. We then discuss the intersection between functional properties of transcription factors and the organization of transcriptional networks. Finally, we present some of the interesting evolutionary conundrums posed by our newly gained understanding of the bacterial transcription apparatus and potential areas for future explorations.
Published by Elsevier Inc.
Figures
Fig. 1
Structure of the bacterial transcription initiation complex. The cartoon representation was derived from an EM structure of the initiation complex (PDB: 3iyd) in association with DNA that contains the α, β, β′, ω, σ70 and the wHTH domains of CRP (CAP) transcription factor. For increased clarity, only the key globular domains of these proteins are shown and labeled. The remaining parts of the structure are shown as coils.
Fig. 2
Structures of key conserved domains of the β, β′ and α subunits. Strands are colored green, whereas helices are colored red or blue. Only the core conserved regions of the domains are shown. Inserts in domains are mostly suppressed or excised as depicted. The C-terminal domain of the ribosomal L25 protein is also depicted to illustrate its structural relationship with the conserved domain inserted into the ASCR domain of the α subunit (L25C–like domain). Structural elements in the L25C–like domain of the α subunit that are not present in the ribosomal L25 protein are colored orange.
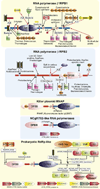
Fig. 3
Domain architectures of the RNA polymerase β and β′ subunits, yeast killer plasmid RNA polymerase, NCgl1702-like RNA polymerases and the prokaryotic RdRP-like RNA polymerases. For the β and β′ subunits, the domain architecture reconstructed to the last universal common ancestor is shown in the center and inserts in various lineages are shown around this core. Archaeo-eukaryotic domain inserts are indicated with a red arrow and bacterial inserts are marked with a black arrow. Lineages in which the inserts are observed are indicated near the arrows or architecture. Red asterisks indicate new domains discovered in this study. Bacterial inserts, on occasions, differ within members of a closely related bacterial lineage. For a more detailed discussion of these variations, refer to Lane and Darst (2010a). A similar representation is used for the prokaryotic RdRP-like proteins, where lineage-specific inserts are marked with a representative gene and species name around a core conserved architecture. Genes in operons are shown as box-arrows with the arrow head pointing from the 5′ to the 3′ direction of the coding sequence. Operons are labeled with the gene name of the polymerase gene and species name. Refer to the supplement for more detailed domain architectures and gene neighborhoods. Standard abbreviations are used for domain and lineage names. The DCL domain is an RNA binding domain which is also found in a stand-alone form in bacteria and in several eukaryotic rRNA biogenesis proteins. Other abbreviations: A, E: archaea and eukaryotes, ASCR: alpha subunit core related, ATL: AT-Hook like motifs, PPI: peptidyl prolyl isomerase, ZnR: zinc ribbon.
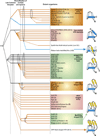
Fig. 4
Higher order evolutionary relationships of bacterial specific transcription factors containing a HTH domain. The horizontal lines represent temporal epochs corresponding to major transitions in evolution of bacteria, namely the last universal common ancestor and the diversification of archaea and bacteria. Solid lines reflect the maximum depth of time to which a particular family can be traced. Broken lines indicate an uncertainty with respect to the exact point of origin of a lineage. The ellipses encompass groups of lineages from which a new lineage with relatively limited distribution could have potentially emerged. Lineages of archaeal origin are colored blue, those of bacterial origin are colored orange and those present in archaea and bacteria are colored black. The phyletic distribution of the lineages are also shown in brackets, where A: Archaea; B: bacteria and E: eukaryotes. The “>” reflects lateral transfer with the arrow head pointing to the potential direction of transfer. Also shown to the right are cartoon representations of the major structural types of HTH domains found in bacterial transcription factors. The TFIIB lineage of archaeo-eukaryotic HTHs is shown to illustrate its relationship with the sigma factor.
Fig. 5
Examples of domain architectures of bacterial transcription factors described in the text. Proteins are labeled with their gene and species names. The domains are not drawn to scale. Standard nomenclatures were mostly used to depict the various domains. Some additional abbreviations include: TM: transmembrane, σ-54 N: globular domain found at the N-terminus of σ54, Sigma-N2 and SigmaN: Conserved N-terminal domains found in σ70, BTAD: conserved domain found in bacterial signaling proteins, ZnRib: Zinc ribbon, FER: classical Ferredoxin domain of the RRM fold.
Fig. 6
Scaling of bacterial transcription factors with proteome size. All graphs show a scatter plot of number of transcription factors in a given proteome (_Y_-axis) versus the number of protein-coding genes in that organism (_X_-axis). In (A) and (B), the _Y_-axis is the overall number of transcription factors across bacteria and in individual lineages respectively. In (C) The _Y_-axis is the number of predicted two-component system proteins. Note that anomalous numbers in Geobacillus and Paenibacillus that are shown as red points. (D) The _Y_-axis is number of one-component system and other phospho-relay system proteins.
Similar articles
- Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases.
Iyer LM, Koonin EV, Aravind L. Iyer LM, et al. BMC Struct Biol. 2003 Jan 28;3:1. doi: 10.1186/1472-6807-3-1. Epub 2003 Jan 28. BMC Struct Biol. 2003. PMID: 12553882 Free PMC article. - Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer.
Iyer LM, Koonin EV, Aravind L. Iyer LM, et al. Gene. 2004 Jun 23;335:73-88. doi: 10.1016/j.gene.2004.03.017. Gene. 2004. PMID: 15194191 - Molecular evolution of multisubunit RNA polymerases: sequence analysis.
Lane WJ, Darst SA. Lane WJ, et al. J Mol Biol. 2010 Jan 29;395(4):671-85. doi: 10.1016/j.jmb.2009.10.062. Epub 2009 Nov 3. J Mol Biol. 2010. PMID: 19895820 Free PMC article. - Evolutionary Origins of Two-Barrel RNA Polymerases and Site-Specific Transcription Initiation.
Fouqueau T, Blombach F, Werner F. Fouqueau T, et al. Annu Rev Microbiol. 2017 Sep 8;71:331-348. doi: 10.1146/annurev-micro-091014-104145. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657884 Review. - The many faces of the helix-turn-helix domain: transcription regulation and beyond.
Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. Aravind L, et al. FEMS Microbiol Rev. 2005 Apr;29(2):231-62. doi: 10.1016/j.femsre.2004.12.008. FEMS Microbiol Rev. 2005. PMID: 15808743 Review.
Cited by
- tRNA evolution from the proto-tRNA minihelix world.
Root-Bernstein R, Kim Y, Sanjay A, Burton ZF. Root-Bernstein R, et al. Transcription. 2016 Oct 19;7(5):153-163. doi: 10.1080/21541264.2016.1235527. Transcription. 2016. PMID: 27636862 Free PMC article. - Basic mechanism of transcription by RNA polymerase II.
Svetlov V, Nudler E. Svetlov V, et al. Biochim Biophys Acta. 2013 Jan;1829(1):20-8. doi: 10.1016/j.bbagrm.2012.08.009. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982365 Free PMC article. Review. - TFIIB is only ∼9 Å away from the 5'-end of a trimeric RNA primer in a functional RNA polymerase II preinitiation complex.
Bick MJ, Malik S, Mustaev A, Darst SA. Bick MJ, et al. PLoS One. 2015 Mar 16;10(3):e0119007. doi: 10.1371/journal.pone.0119007. eCollection 2015. PLoS One. 2015. PMID: 25774659 Free PMC article. - The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems.
Oberstaller J, Pumpalova Y, Schieler A, Llinás M, Kissinger JC. Oberstaller J, et al. Nucleic Acids Res. 2014 Jul;42(13):8271-84. doi: 10.1093/nar/gku500. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957599 Free PMC article. - Rewiring of growth-dependent transcription regulation by a point mutation in region 1.1 of the housekeeping σ factor.
Pletnev P, Pupov D, Pshanichnaya L, Esyunina D, Petushkov I, Nesterchuk M, Osterman I, Rubtsova M, Mardanov A, Ravin N, Sergiev P, Kulbachinskiy A, Dontsova O. Pletnev P, et al. Nucleic Acids Res. 2020 Nov 4;48(19):10802-10819. doi: 10.1093/nar/gkaa798. Nucleic Acids Res. 2020. PMID: 32997144 Free PMC article.
References
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. - PubMed
- Anantharaman V, Aravind L. Diversification of catalytic activities and ligand interactions in the protein fold shared by the sugar isomerases, eIF2B, DeoR transcription factors, acyl-CoA transferases and methenyltetrahydrofolate synthetase. J. Mol. Biol. 2006;356:823–842. - PubMed
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Z99 CL999999/ImNIH/Intramural NIH HHS/United States
- Z99 HL999999/ImNIH/Intramural NIH HHS/United States
- Z99 LM999999/ImNIH/Intramural NIH HHS/United States
- Z99 MH999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources