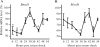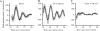A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle - PubMed (original) (raw)
A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle
Xiping Zhang et al. Nucleic Acids Res. 2012 Apr.
Abstract
The myogenic differentiation 1 (MyoD) gene is a master regulator of myogenesis. We previously reported that the expression of MyoD mRNA oscillates over 24 h in skeletal muscle and that the circadian clock transcription factors, BMAL1 (brain and muscle ARNT-like 1) and CLOCK (circadian locomotor output cycles kaput), were bound to the core enhancer (CE) of the MyoD gene in vivo. In this study, we provide in vivo and in vitro evidence that the CE is necessary for circadian expression of MyoD in adult muscle. Gel shift assays identified a conserved non-canonical E-box within the CE that is bound by CLOCK and BMAL1. Functional analysis revealed that this E-box was required for full activation by BMAL1/CLOCK and for in vitro circadian oscillation. Expression profiling of muscle of CE(loxP/loxP) mice found approximately 1300 genes mis-expressed relative to wild-type. Based on the informatics results, we analyzed the respiratory function of mitochondria isolated from wild-type and CE(loxP/loxP) mice. These assays determined that State 5 respiration was significantly reduced in CE(loxP/loxP) muscle. The results of this work identify a novel element in the MyoD enhancer that confers circadian regulation to MyoD in skeletal muscle and suggest that loss of circadian regulation leads to changes in myogenic expression and downstream mitochondrial function.
Figures
Figure 1.
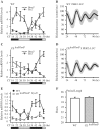
Circadian oscillation of MyoD mRNA is dampened in skeletal muscle tissues of the CEloxP/loxP mouse. Adult C57BL/6 J and CEloxP/loxP mice were entrained to 12 h light/12 h dark conditions for 2 weeks. Thirty hours after lights were turned off (CT18), TA muscles were collected every 4 h from CT18 to CT50. Transcript levels were determined by RT–PCR. Bmal1, Per2 and MyoD mRNA levels were normalized to Rpl26 mRNA levels. (A) Bmal1 and Per2 expression in wild-type mice TA muscle; (B) Luminescence recording from Per2::Luc soleus muscles. The data for the graphs represent the mean values ± SEM of raw data for five animals. (C) Bmal1 and Per2 expression in CEloxP/loxP mice TA muscle; (D) Luminescence recordings from Per2::Luc × CEloxP/loxP soleus muscles. The data for the graphs represent the mean values ± SEM of raw data for five animals. (E) MyoD expression in wild-type and CEloxP/loxP mice TA muscle; (F) Average period length data of bioluminescence from Per2::Luc and Per2::Luc × CEloxP/loxP soleus muscles. The graphs in (A, C and E) display the results as the mean value ± SEM for three independent samples for each genotype. The gray bars indicate the subjective circadian light period and the black bars indicate subjective circadian dark period. For each respective gene, one-way ANOVA followed by Tukey's post-hoc test was performed to determine significant differences across all time points. For circadian analysis of gene expression, the JTK_CYCLE program was used.
Figure 2.
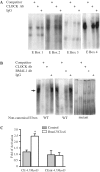
The non-canonical E-box of the MyoD CE is required for CLOCK and BMAL1 binding and activation of the MyoD promoter by BMAL1: CLOCK. EMSA analysis of CLOCK and BMAL1 binding at the E-boxes located within the MyoD CE was performed with nuclear extract derived from C2C12 cells over-expressing Clock and Bmal1 and an oligonucleotide probe containing one of the five CE E-boxes. (A) The DNA:protein complexes formed using a probe containing one of the canonical E-boxes (E1, E2, E3 or E4). (B) The DNA:protein complexes formed using the non-canonical E-box and the mutant of the non-canonical E-box. The arrow indicates the specific DNA:protein complex that contained CLOCK and BMAL1. (C) A wild-type (CE-4.7_MyoD_:luciferase) or mutant (CEmt-4.7_MyoD_:luciferase) reporter gene construct was transiently co-transfected with Clock plus Bmal1 expression plasmids into C2C12 myoblasts. Luciferase activity was measured 24–48 h later. The histogram displays the results from three independent experiments with values presented as mean ± SEM. A one-way ANOVA test indicated CE-4.7_MyoD_:luciferase reporter gene activation by CLOCK plus BMAL1 (empty bar) was significantly greater than control (gray bar) denoted by asterisks.
Figure 3.
Serum shock is sufficient to synchronize circadian gene expression in C2C12 myotubes. Stable clones of C2C12 cells (CE-4.7_MyoD_) were differentiated for 5 days prior to serum shock. RNA was isolated at the indicated time post-serum shock and used to determine the expression level of Bmal1 and MyoD by RT-PCR. The target transcript levels were normalized to Rpl26 transcript levels and then normalized to the highest point across all time points. The data for the graphs represent the mean values ± SEM for three independent clones for the reporter gene. The expression of endogenous Bmal1 (A), endogenous MyoD (B) all displayed a circadian oscillation as indicated by significant differences in peak and trough expression. For each respective gene, one-way ANOVA followed by Tukey post-hoc test identified significant difference (P < 0.05) between peak expression and other time points as indicated by an asterisk. For circadian analysis of gene expression the JTK_CYCLE program was used.
Figure 4.
Mutation of the MyoD CE non-canonical E-box disrupts circadian oscillation in bioluminescent assay. Stable clones of C2C12 cells (carrying the Bmal1 promoter:luciferase, CE-4.7_MyoD_:luciferase or CEmt-4.7_MyoD_:luciferase reporter) were differentiated for 5 days prior to serum shock. Then the cultures were incubated in Lumicycle at 37°C and bioluminescence was recorded. The graphs shown here are baseline subtracted results as the mean value ± SEM from three independent clones for the reporter gene. (A) Bmal1 promoter:luciferase reporter. (B) CE-4.7_MyoD_:luciferase reporter. (C) CEmt-4.7_MyoD_:luciferase reporter. The graphs display the results as the mean value ± SEM from three independent clones for each reporter. For circadian analysis of gene expression, the JTK_CYCLE program was used.
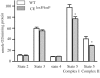
Figure 5.
Respiration rates are lower in the mitochondria of the CEloxP/loxP than in wild-type. Gastrocnemius muscles from 5- to 6-month-old mice were isolated from wild-type (empty bars) or the CEloxP/loxP (gray bars) and used for the determination of respiration rates. The data for the graphs represent the mean values ± SEM for four animals.
Similar articles
- CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function.
Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Andrews JL, et al. Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):19090-5. doi: 10.1073/pnas.1014523107. Epub 2010 Oct 18. Proc Natl Acad Sci U S A. 2010. PMID: 20956306 Free PMC article. - MYOD1 functions as a clock amplifier as well as a critical co-factor for downstream circadian gene expression in muscle.
Hodge BA, Zhang X, Gutierrez-Monreal MA, Cao Y, Hammers DW, Yao Z, Wolff CA, Du P, Kemler D, Judge AR, Esser KA. Hodge BA, et al. Elife. 2019 Feb 21;8:e43017. doi: 10.7554/eLife.43017. Elife. 2019. PMID: 30789342 Free PMC article. - CLOCK and BMAL1 Regulate Muscle Insulin Sensitivity via SIRT1 in Male Mice.
Liu J, Zhou B, Yan M, Huang R, Wang Y, He Z, Yang Y, Dai C, Wang Y, Zhang F, Zhai Q. Liu J, et al. Endocrinology. 2016 Jun;157(6):2259-69. doi: 10.1210/en.2015-2027. Epub 2016 Apr 1. Endocrinology. 2016. PMID: 27035655 - Circadian rhythms, the molecular clock, and skeletal muscle.
Harfmann BD, Schroder EA, Esser KA. Harfmann BD, et al. J Biol Rhythms. 2015 Apr;30(2):84-94. doi: 10.1177/0748730414561638. Epub 2014 Dec 15. J Biol Rhythms. 2015. PMID: 25512305 Free PMC article. Review. - Circadian rhythms, the molecular clock, and skeletal muscle.
Lefta M, Wolff G, Esser KA. Lefta M, et al. Curr Top Dev Biol. 2011;96:231-71. doi: 10.1016/B978-0-12-385940-2.00009-7. Curr Top Dev Biol. 2011. PMID: 21621073 Free PMC article. Review.
Cited by
- Circadian clock regulation of skeletal muscle growth and repair.
Chatterjee S, Ma K. Chatterjee S, et al. F1000Res. 2016 Jun 30;5:1549. doi: 10.12688/f1000research.9076.1. eCollection 2016. F1000Res. 2016. PMID: 27540471 Free PMC article. Review. - BMAL1 drives muscle repair through control of hypoxic NAD+ regeneration in satellite cells.
Zhu P, Hamlish NX, Thakkar AV, Steffeck AWT, Rendleman EJ, Khan NH, Waldeck NJ, DeVilbiss AW, Martin-Sandoval MS, Mathews TP, Chandel NS, Peek CB. Zhu P, et al. Genes Dev. 2022 Feb 1;36(3-4):149-166. doi: 10.1101/gad.349066.121. Epub 2022 Feb 3. Genes Dev. 2022. PMID: 35115380 Free PMC article. - The circadian E3 ligase FBXL21 regulates myoblast differentiation and sarcomere architecture via MYOZ1 ubiquitination and NFAT signaling.
Lim JY, Kim E, Douglas CM, Wirianto M, Han C, Ono K, Kim SY, Ji JH, Tran CK, Chen Z, Esser KA, Yoo SH. Lim JY, et al. PLoS Genet. 2022 Dec 27;18(12):e1010574. doi: 10.1371/journal.pgen.1010574. eCollection 2022 Dec. PLoS Genet. 2022. PMID: 36574402 Free PMC article. - Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice.
Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, Bass J. Peek CB, et al. Science. 2013 Nov 1;342(6158):1243417. doi: 10.1126/science.1243417. Epub 2013 Sep 19. Science. 2013. PMID: 24051248 Free PMC article. - The GSK-3β-FBXL21 Axis Contributes to Circadian TCAP Degradation and Skeletal Muscle Function.
Wirianto M, Yang J, Kim E, Gao S, Paudel KR, Choi JM, Choe J, Gloston GF, Ademoji P, Parakramaweera R, Jin J, Esser KA, Jung SY, Geng YJ, Lee HK, Chen Z, Yoo SH. Wirianto M, et al. Cell Rep. 2020 Sep 15;32(11):108140. doi: 10.1016/j.celrep.2020.108140. Cell Rep. 2020. PMID: 32937135 Free PMC article.
References
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. - PubMed
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. - PubMed
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases